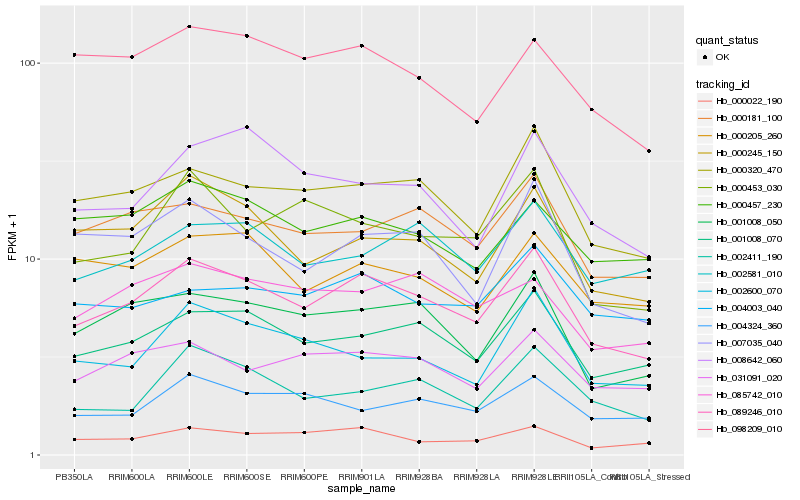
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089246_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_24395 [Jatropha curcas] |
| 2 |
Hb_001008_070 |
0.0736282768 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 3 |
Hb_000320_470 |
0.075904175 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 4 |
Hb_001008_050 |
0.086102671 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 5 |
Hb_000181_100 |
0.0906976421 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 6 |
Hb_031091_020 |
0.0926536906 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 7 |
Hb_000022_190 |
0.0937692569 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22690-like [Jatropha curcas] |
| 8 |
Hb_000245_150 |
0.0947447082 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FtsH [Jatropha curcas] |
| 9 |
Hb_007035_040 |
0.0954363538 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_085742_010 |
0.1038888813 |
- |
- |
hypothetical protein POPTR_0005s05170g [Populus trichocarpa] |
| 11 |
Hb_002581_010 |
0.1040220596 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 12 |
Hb_098209_010 |
0.1041213105 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 13 |
Hb_002411_190 |
0.1043901209 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 14 |
Hb_000453_030 |
0.1045869703 |
- |
- |
Protein kinase capable of phosphorylating tyrosine family protein [Populus trichocarpa] |
| 15 |
Hb_000205_260 |
0.1052233271 |
transcription factor |
TF Family: SNF2 |
hypothetical protein JCGZ_23567 [Jatropha curcas] |
| 16 |
Hb_002600_070 |
0.1058989445 |
transcription factor |
TF Family: TAZ |
PREDICTED: histone acetyltransferase HAC12 [Jatropha curcas] |
| 17 |
Hb_004324_360 |
0.1059220426 |
- |
- |
PREDICTED: uncharacterized protein LOC105650600 isoform X1 [Jatropha curcas] |
| 18 |
Hb_008642_060 |
0.1068550155 |
- |
- |
PREDICTED: dynamin-related protein 1E isoform X2 [Jatropha curcas] |
| 19 |
Hb_000457_230 |
0.1071522222 |
- |
- |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_004003_040 |
0.1075289648 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |