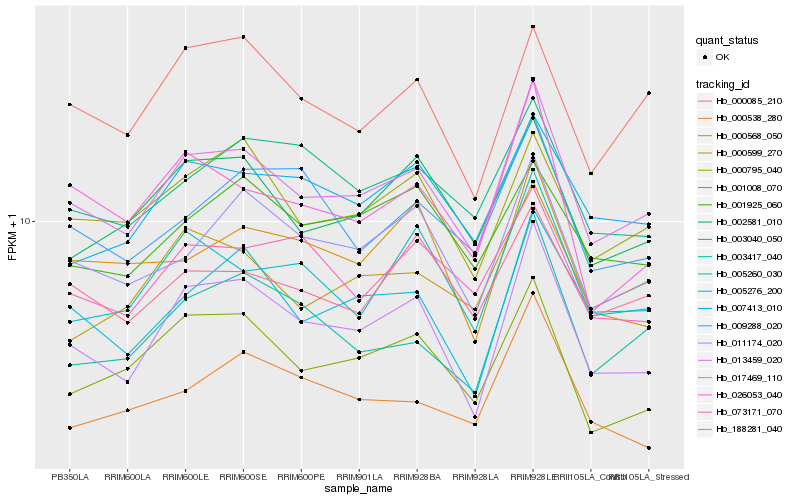
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_017469_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 2 |
Hb_013459_020 |
0.0495022791 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 3 |
Hb_011174_020 |
0.0591687489 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 4 |
Hb_000795_040 |
0.0607234657 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 5 |
Hb_005276_200 |
0.0643062305 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002581_010 |
0.0647108085 |
- |
- |
PREDICTED: uncharacterized protein LOC105641469 isoform X2 [Jatropha curcas] |
| 7 |
Hb_001008_070 |
0.0663527403 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 8 |
Hb_188281_040 |
0.0667692319 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 9 |
Hb_003417_040 |
0.0676736606 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 10 |
Hb_000599_270 |
0.0683763698 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 11 |
Hb_000085_210 |
0.0696014611 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 12 |
Hb_005260_030 |
0.0702736586 |
- |
- |
PREDICTED: uncharacterized protein LOC105633782 [Jatropha curcas] |
| 13 |
Hb_003040_050 |
0.0705574836 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 14 |
Hb_007413_010 |
0.0712581325 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 15 |
Hb_026053_040 |
0.0714191593 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_073171_070 |
0.0718273492 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 17 |
Hb_001925_060 |
0.0740248087 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 18 |
Hb_009288_020 |
0.0748933002 |
- |
- |
PREDICTED: protein FRIGIDA [Jatropha curcas] |
| 19 |
Hb_000538_280 |
0.0791726714 |
- |
- |
PREDICTED: uncharacterized protein LOC105642112 [Jatropha curcas] |
| 20 |
Hb_000568_050 |
0.0799272437 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |