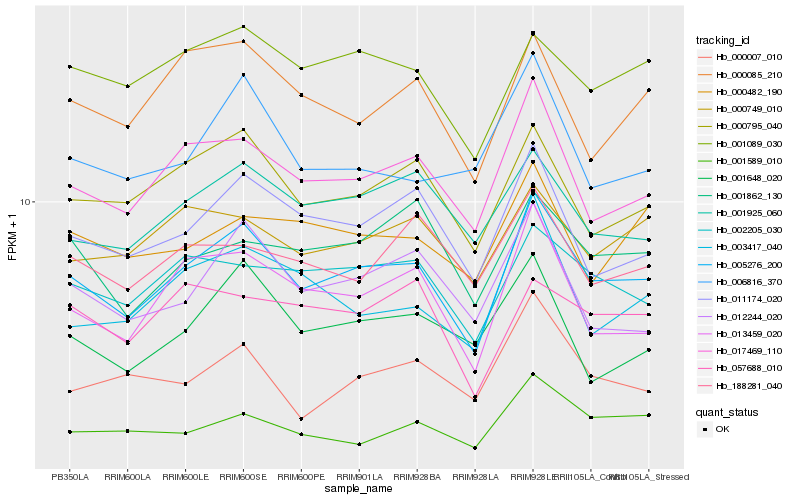
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005276_200 |
0.0 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_017469_110 |
0.0643062305 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 3 |
Hb_000795_040 |
0.0734538685 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 4 |
Hb_011174_020 |
0.0747588244 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 5 |
Hb_013459_020 |
0.075782512 |
- |
- |
PREDICTED: uncharacterized protein LOC105628886 [Jatropha curcas] |
| 6 |
Hb_002205_030 |
0.0791399366 |
- |
- |
PREDICTED: alpha-galactosidase 3 [Jatropha curcas] |
| 7 |
Hb_001925_060 |
0.0841469623 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |
| 8 |
Hb_001089_030 |
0.0862171716 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 9 |
Hb_000085_210 |
0.0862743083 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 10 |
Hb_012244_020 |
0.0873654893 |
- |
- |
calpain, putative [Ricinus communis] |
| 11 |
Hb_000749_010 |
0.0902235302 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_003417_040 |
0.0905479967 |
transcription factor |
TF Family: mTERF |
hypothetical protein JCGZ_10155 [Jatropha curcas] |
| 13 |
Hb_001589_010 |
0.0914046457 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 14 |
Hb_001862_130 |
0.092100406 |
- |
- |
Protein FRIGIDA, putative [Ricinus communis] |
| 15 |
Hb_001648_020 |
0.0941891185 |
- |
- |
hypothetical protein POPTR_0007s07750g [Populus trichocarpa] |
| 16 |
Hb_188281_040 |
0.0951202245 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 17 |
Hb_000007_010 |
0.095191965 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 18 |
Hb_057688_010 |
0.0952395129 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 isoform X2 [Jatropha curcas] |
| 19 |
Hb_006816_370 |
0.0953742473 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105647456 [Jatropha curcas] |
| 20 |
Hb_000482_190 |
0.0957057797 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |