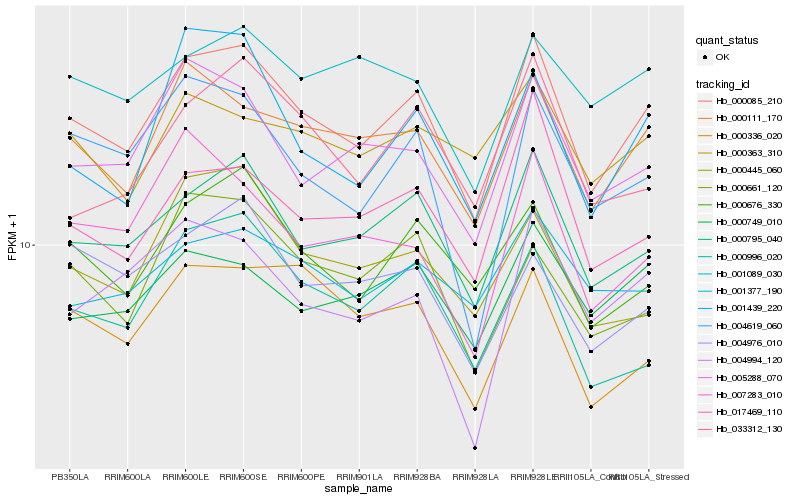
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000085_210 |
0.0 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 2 |
Hb_000795_040 |
0.0503472261 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 3 |
Hb_000111_170 |
0.0554585708 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005288_070 |
0.0677548451 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 5 |
Hb_000676_330 |
0.0682304459 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_017469_110 |
0.0696014611 |
- |
- |
PREDICTED: uncharacterized protein LOC105641795 [Jatropha curcas] |
| 7 |
Hb_001439_220 |
0.0722179404 |
- |
- |
xenotropic and polytropic murine leukemia virus receptor ids-4, putative [Ricinus communis] |
| 8 |
Hb_000749_010 |
0.0724045316 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 9 |
Hb_004619_060 |
0.0761134994 |
- |
- |
PREDICTED: uncharacterized protein At1g10890-like [Nelumbo nucifera] |
| 10 |
Hb_000661_120 |
0.0763575793 |
- |
- |
cap binding protein, putative [Ricinus communis] |
| 11 |
Hb_000996_020 |
0.0777739762 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 12 |
Hb_001377_190 |
0.078909373 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000336_020 |
0.0790952701 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 14 |
Hb_001089_030 |
0.0791136381 |
- |
- |
PREDICTED: uncharacterized protein LOC105638026 [Jatropha curcas] |
| 15 |
Hb_000445_060 |
0.0792252837 |
- |
- |
PREDICTED: uncharacterized protein LOC105638996 isoform X3 [Jatropha curcas] |
| 16 |
Hb_004994_120 |
0.0797825476 |
- |
- |
PREDICTED: acyl-coenzyme A thioesterase 13-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_033312_130 |
0.080539995 |
- |
- |
PREDICTED: protein TIC 40, chloroplastic [Jatropha curcas] |
| 18 |
Hb_007283_010 |
0.0811353102 |
- |
- |
PREDICTED: pre-mRNA-processing protein 40C isoform X1 [Jatropha curcas] |
| 19 |
Hb_004976_010 |
0.0816966662 |
- |
- |
hypothetical protein RCOM_1429110 [Ricinus communis] |
| 20 |
Hb_000363_310 |
0.082208437 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |