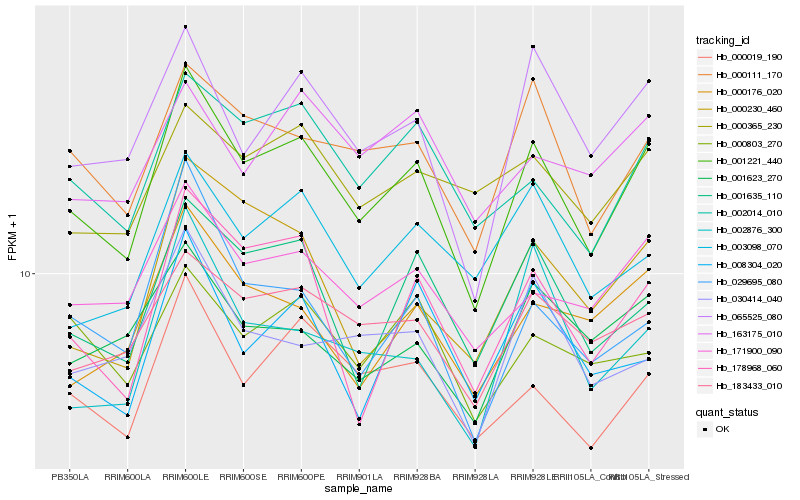
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001221_440 |
0.0 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000111_170 |
0.06389163 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 3 |
Hb_065525_080 |
0.0684168565 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 14 [Populus euphratica] |
| 4 |
Hb_000019_190 |
0.0687635437 |
- |
- |
hypothetical protein POPTR_0002s23900g [Populus trichocarpa] |
| 5 |
Hb_001635_110 |
0.0689155823 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 6 |
Hb_171900_090 |
0.0689214098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002014_010 |
0.0692508141 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 8 |
Hb_178968_060 |
0.071366461 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 9 |
Hb_183433_010 |
0.0720893508 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 10 |
Hb_029695_080 |
0.0734319683 |
- |
- |
PREDICTED: NAD-dependent protein deacetylase SRT1 [Prunus mume] |
| 11 |
Hb_000176_020 |
0.0777639272 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |
| 12 |
Hb_000230_460 |
0.0803518115 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 13 |
Hb_001623_270 |
0.0804262904 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 14 |
Hb_003098_070 |
0.0814953018 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 15 |
Hb_008304_020 |
0.0817464246 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 16 |
Hb_030414_040 |
0.0817944014 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002876_300 |
0.0818759728 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_000803_270 |
0.0833622018 |
- |
- |
PREDICTED: nuclear cap-binding protein subunit 1 [Jatropha curcas] |
| 19 |
Hb_163175_010 |
0.0857143374 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 20 |
Hb_000365_230 |
0.0859113028 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |