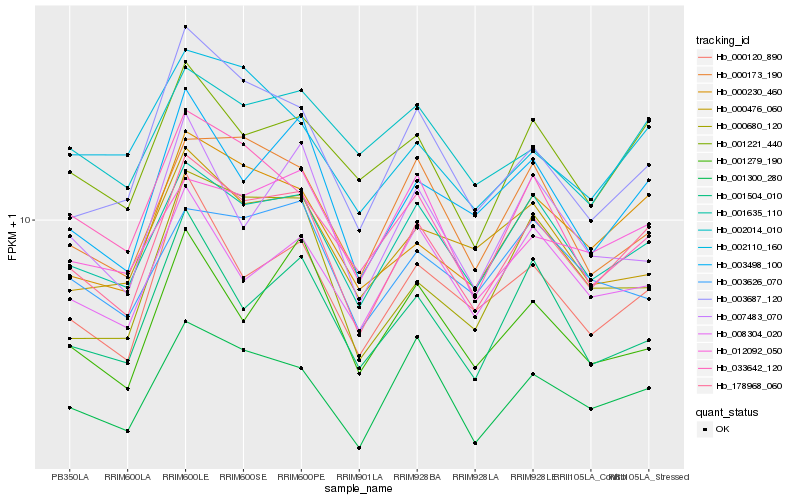
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_178968_060 |
0.0 |
- |
- |
PREDICTED: ubiquitin receptor RAD23b-like [Gossypium raimondii] |
| 2 |
Hb_001635_110 |
0.0486705232 |
- |
- |
PREDICTED: NADP-specific glutamate dehydrogenase [Jatropha curcas] |
| 3 |
Hb_000230_460 |
0.0631502841 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 4 |
Hb_001300_280 |
0.0642095781 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein ucpB [Jatropha curcas] |
| 5 |
Hb_003498_100 |
0.064709908 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 6 |
Hb_000120_890 |
0.0701998019 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 7 |
Hb_001221_440 |
0.071366461 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000173_190 |
0.0751039093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_003687_120 |
0.0803774561 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 10 |
Hb_033642_120 |
0.082195378 |
- |
- |
PREDICTED: nucleolar GTP-binding protein 2 [Vitis vinifera] |
| 11 |
Hb_008304_020 |
0.0830655858 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 12 |
Hb_001504_010 |
0.0839305528 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 13 |
Hb_012092_050 |
0.0844091177 |
- |
- |
PREDICTED: polypyrimidine tract-binding protein homolog 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_003626_070 |
0.0845883464 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 15 |
Hb_000476_060 |
0.0856019882 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 16 |
Hb_000680_120 |
0.0873372448 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 17 |
Hb_007483_070 |
0.0883244794 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 18 |
Hb_002110_160 |
0.0887716616 |
- |
- |
PREDICTED: phosphoribosylaminoimidazole-succinocarboxamide synthase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002014_010 |
0.089374508 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_001279_190 |
0.089559843 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |