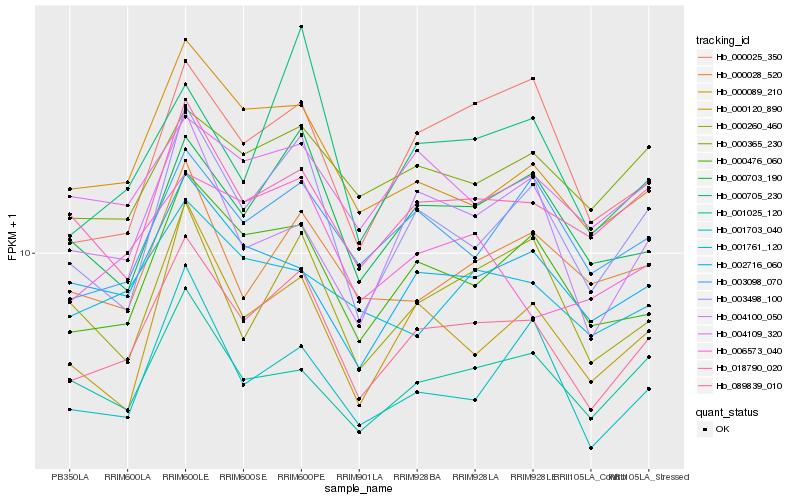
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089839_010 |
0.0 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 2 |
Hb_000476_060 |
0.0611140052 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 3 |
Hb_003498_100 |
0.0678324188 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 4 |
Hb_000703_190 |
0.0710356219 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_018790_020 |
0.0749097536 |
- |
- |
protein kinase, putative [Ricinus communis] |
| 6 |
Hb_000089_210 |
0.0750853128 |
- |
- |
unknown [Medicago truncatula] |
| 7 |
Hb_003098_070 |
0.0769991611 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 8 |
Hb_000120_890 |
0.0773587083 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 9 |
Hb_002716_060 |
0.0795454817 |
- |
- |
carboxylesterase np, putative [Ricinus communis] |
| 10 |
Hb_001703_040 |
0.0805185792 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 11 |
Hb_004109_320 |
0.0805628407 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 52 A [Jatropha curcas] |
| 12 |
Hb_000365_230 |
0.0814180585 |
- |
- |
PREDICTED: uncharacterized protein LOC105649056 [Jatropha curcas] |
| 13 |
Hb_001025_120 |
0.0821195319 |
transcription factor |
TF Family: Jumonji |
PREDICTED: jmjC domain-containing protein 4 [Jatropha curcas] |
| 14 |
Hb_000028_520 |
0.0846055994 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 15 |
Hb_000260_460 |
0.0858256834 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000025_350 |
0.0873360668 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000705_230 |
0.0880835364 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 18 |
Hb_006573_040 |
0.0884271851 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 19 |
Hb_001761_120 |
0.0891640362 |
- |
- |
hypothetical protein JCGZ_22564 [Jatropha curcas] |
| 20 |
Hb_004100_050 |
0.0894060953 |
- |
- |
PREDICTED: mitochondrial-processing peptidase subunit alpha-like [Jatropha curcas] |