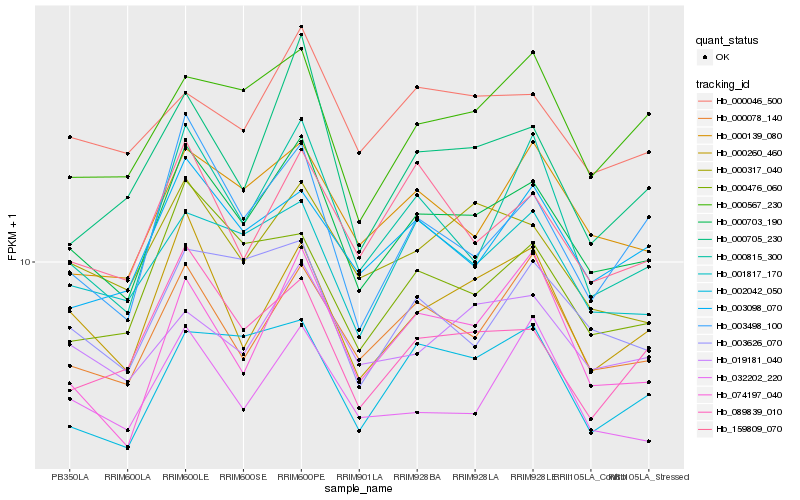
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000703_190 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000260_460 |
0.0614051854 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000078_140 |
0.0635932387 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 4 |
Hb_001817_170 |
0.0648982786 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 5 |
Hb_000139_080 |
0.0668076646 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000317_040 |
0.0673385176 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 7 |
Hb_159809_070 |
0.0704245692 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 8 |
Hb_089839_010 |
0.0710356219 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 9 |
Hb_000815_300 |
0.0710517308 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 10 |
Hb_000476_060 |
0.0711308881 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_003626_070 |
0.0717576715 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 12 |
Hb_002042_050 |
0.072097103 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003098_070 |
0.0726843733 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 14 |
Hb_000046_500 |
0.0730508478 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 15 |
Hb_003498_100 |
0.0732736212 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 16 |
Hb_019181_040 |
0.0735187722 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 17 |
Hb_000567_230 |
0.0739319234 |
- |
- |
PREDICTED: peroxisome biogenesis protein 7 [Jatropha curcas] |
| 18 |
Hb_032202_220 |
0.0755770264 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_000705_230 |
0.0765132824 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1 isozyme 2 [Jatropha curcas] |
| 20 |
Hb_074197_040 |
0.0769478318 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |