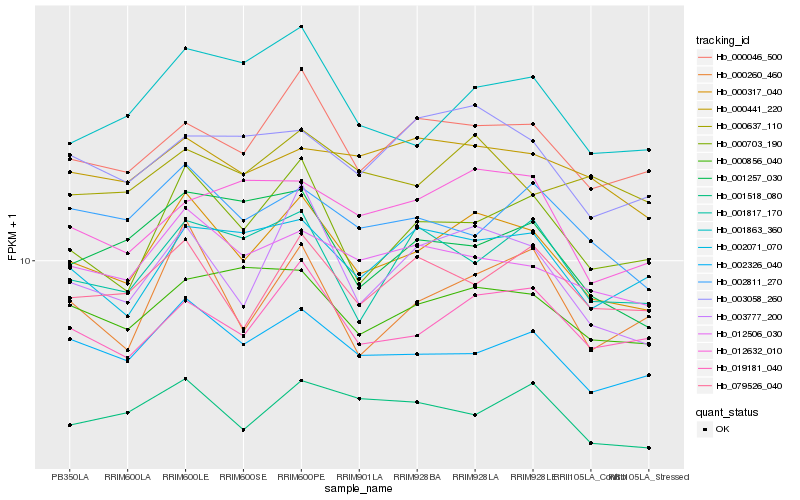
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_040 |
0.0 |
- |
- |
PREDICTED: xylosyltransferase 1-like [Jatropha curcas] |
| 2 |
Hb_003777_200 |
0.063917127 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 3 |
Hb_000703_190 |
0.0673385176 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_019181_040 |
0.0685335709 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 5 |
Hb_002326_040 |
0.0686739161 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 6 |
Hb_000046_500 |
0.0710106842 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Jatropha curcas] |
| 7 |
Hb_012506_030 |
0.0714998724 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 8 |
Hb_000260_460 |
0.0793548916 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 9 |
Hb_000856_040 |
0.079776379 |
transcription factor |
TF Family: DDT |
PREDICTED: uncharacterized protein LOC105640470 [Jatropha curcas] |
| 10 |
Hb_079526_040 |
0.0822138057 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 11 |
Hb_001518_080 |
0.0823804519 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 12 |
Hb_002071_070 |
0.0825440496 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001863_360 |
0.0832217636 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase-like 4 [Jatropha curcas] |
| 14 |
Hb_003058_260 |
0.0848094823 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 15 |
Hb_002811_270 |
0.0857701842 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 16 |
Hb_012632_010 |
0.0880671672 |
- |
- |
PREDICTED: uncharacterized protein LOC105640019 [Jatropha curcas] |
| 17 |
Hb_001817_170 |
0.0881588383 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 18 |
Hb_000441_220 |
0.0889285721 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_001257_030 |
0.0890656747 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek5 [Jatropha curcas] |
| 20 |
Hb_000637_110 |
0.0891156808 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |