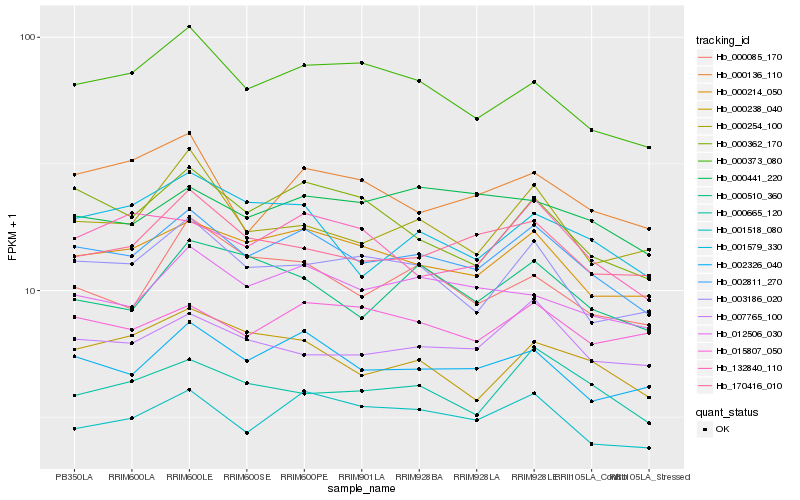
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_270 |
0.0 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 2 |
Hb_000214_050 |
0.0502714184 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 3 |
Hb_002326_040 |
0.053949241 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 4 |
Hb_001518_080 |
0.0593555899 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 5 |
Hb_012506_030 |
0.0635781219 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 6 |
Hb_000373_080 |
0.0655943769 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 7 |
Hb_003186_020 |
0.0665355722 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000362_170 |
0.0686978707 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 9 |
Hb_170416_010 |
0.0688230273 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 10 |
Hb_000665_120 |
0.0708871046 |
transcription factor |
TF Family: SET |
PREDICTED: uncharacterized protein LOC105637593 [Jatropha curcas] |
| 11 |
Hb_007765_100 |
0.0713199617 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 12 |
Hb_000238_040 |
0.0721249706 |
- |
- |
PREDICTED: uncharacterized protein LOC102611758 [Citrus sinensis] |
| 13 |
Hb_001579_330 |
0.0723053723 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X2 [Jatropha curcas] |
| 14 |
Hb_000085_170 |
0.075118276 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_015807_050 |
0.0752534103 |
- |
- |
PREDICTED: uncharacterized protein LOC105632878 [Jatropha curcas] |
| 16 |
Hb_132840_110 |
0.0753258444 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_000441_220 |
0.0754207619 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000510_360 |
0.0757053328 |
transcription factor |
TF Family: GNAT |
PREDICTED: histone acetyltransferase GCN5 [Jatropha curcas] |
| 19 |
Hb_000254_100 |
0.0757629103 |
- |
- |
PREDICTED: glyoxylate/hydroxypyruvate reductase A HPR2 [Jatropha curcas] |
| 20 |
Hb_000136_110 |
0.0769306364 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |