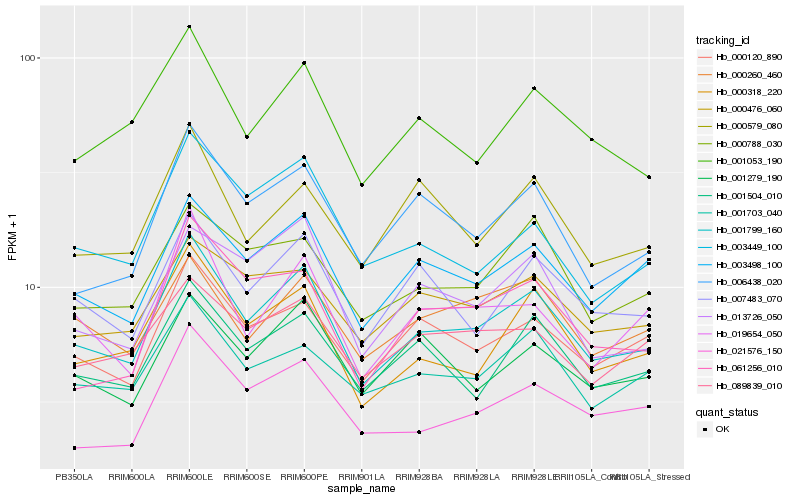
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001799_160 |
0.0 |
- |
- |
PREDICTED: protein VASCULAR ASSOCIATED DEATH 1, chloroplastic isoform X2 [Jatropha curcas] |
| 2 |
Hb_000318_220 |
0.0708671699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003498_100 |
0.0734837999 |
- |
- |
component of oligomeric golgi complex, putative [Ricinus communis] |
| 4 |
Hb_000120_890 |
0.0781377866 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 5 |
Hb_001504_010 |
0.0810143613 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 6 |
Hb_007483_070 |
0.0826640948 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 7 |
Hb_001053_190 |
0.0831043662 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 8 |
Hb_001703_040 |
0.0839809358 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 9 |
Hb_006438_020 |
0.0847875385 |
- |
- |
PREDICTED: lysine--tRNA ligase-like [Populus euphratica] |
| 10 |
Hb_021576_150 |
0.0854788693 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 11 |
Hb_000788_030 |
0.0863983826 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 12 |
Hb_019654_050 |
0.0872686721 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 13 |
Hb_000476_060 |
0.0908449588 |
- |
- |
Tetratricopeptide repeat (TPR)-like superfamily protein isoform 1 [Theobroma cacao] |
| 14 |
Hb_000260_460 |
0.0916505518 |
- |
- |
Ethanolamine-phosphate cytidylyltransferase, putative [Ricinus communis] |
| 15 |
Hb_001279_190 |
0.0921473431 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003449_100 |
0.0925046653 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 17 |
Hb_089839_010 |
0.0939095926 |
- |
- |
PREDICTED: probable xyloglucan glycosyltransferase 6 [Jatropha curcas] |
| 18 |
Hb_061256_010 |
0.0941787688 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 19 |
Hb_013726_050 |
0.0943644257 |
- |
- |
PREDICTED: probable methyltransferase PMT13 [Jatropha curcas] |
| 20 |
Hb_000579_080 |
0.0946314997 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |