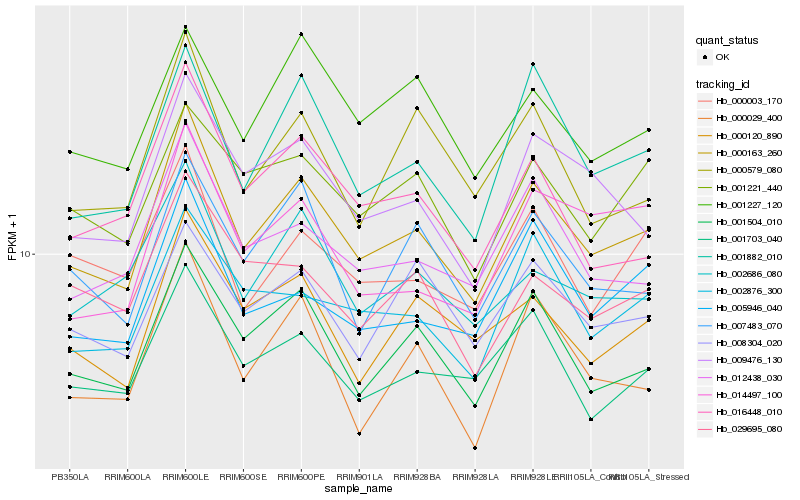
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000163_260 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 2 |
Hb_001504_010 |
0.0632376045 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 3 |
Hb_001882_010 |
0.0718985689 |
- |
- |
- |
| 4 |
Hb_012438_030 |
0.0722686039 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 5 |
Hb_009476_130 |
0.0744143619 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 6 |
Hb_001703_040 |
0.0758261767 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000120_890 |
0.0778157798 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC6 [Jatropha curcas] |
| 8 |
Hb_002686_080 |
0.0801118512 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein ING2-like [Jatropha curcas] |
| 9 |
Hb_016448_010 |
0.0812099737 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 10 |
Hb_005946_040 |
0.0829637495 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 11 |
Hb_007483_070 |
0.0830027709 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 12 |
Hb_002876_300 |
0.0830315449 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000029_400 |
0.0830514678 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000003_170 |
0.0844209688 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 15 |
Hb_000579_080 |
0.08596875 |
- |
- |
PREDICTED: 26S protease regulatory subunit S10B homolog B [Jatropha curcas] |
| 16 |
Hb_001221_440 |
0.0866853483 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 1B-1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_008304_020 |
0.0871414963 |
- |
- |
PREDICTED: ATP-dependent RNA helicase DBP2-like [Jatropha curcas] |
| 18 |
Hb_001227_120 |
0.0875515088 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 19 |
Hb_029695_080 |
0.0882998315 |
- |
- |
PREDICTED: NAD-dependent protein deacetylase SRT1 [Prunus mume] |
| 20 |
Hb_014497_100 |
0.0902239779 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |