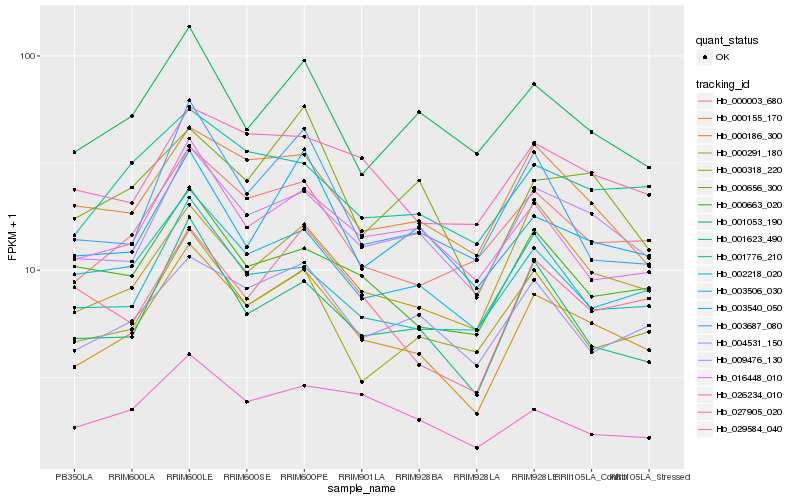
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_300 |
0.0 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_009476_130 |
0.0845000415 |
- |
- |
hypothetical protein VITISV_005430 [Vitis vinifera] |
| 3 |
Hb_000003_680 |
0.0987337726 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 4 |
Hb_003506_030 |
0.1038542143 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 5 |
Hb_001623_490 |
0.1049755216 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000291_180 |
0.1052714806 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 7 |
Hb_004531_150 |
0.1060990544 |
- |
- |
PREDICTED: peroxisome biogenesis protein 2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000155_170 |
0.1096227378 |
transcription factor |
TF Family: VOZ |
hypothetical protein EUTSA_v10007498mg [Eutrema salsugineum] |
| 9 |
Hb_001776_210 |
0.1129129497 |
- |
- |
CBL-interacting serine/threonine-protein kinase, putative [Ricinus communis] |
| 10 |
Hb_003540_050 |
0.1130060638 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 11 |
Hb_016448_010 |
0.1130405587 |
- |
- |
Protein MYG1, putative [Ricinus communis] |
| 12 |
Hb_027905_020 |
0.1154738453 |
- |
- |
PREDICTED: uncharacterized protein LOC105642268 [Jatropha curcas] |
| 13 |
Hb_002218_020 |
0.1160833996 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 14 |
Hb_000656_300 |
0.1166254941 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 15 |
Hb_029584_040 |
0.1171692879 |
- |
- |
hypothetical protein Csa_1G586790 [Cucumis sativus] |
| 16 |
Hb_003687_080 |
0.1182064396 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 17 |
Hb_026234_010 |
0.1182612205 |
- |
- |
choline monooxygenase, putative [Ricinus communis] |
| 18 |
Hb_000663_020 |
0.1182753648 |
- |
- |
PREDICTED: alternative NAD(P)H-ubiquinone oxidoreductase C1, chloroplastic/mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_001053_190 |
0.1183627696 |
- |
- |
ADP,ATP carrier protein, putative [Ricinus communis] |
| 20 |
Hb_000318_220 |
0.1184436734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |