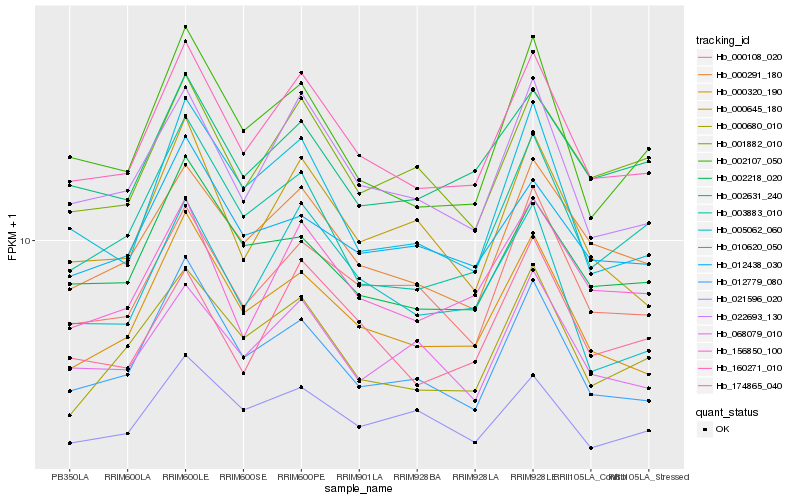
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_160271_010 |
0.0 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_022693_130 |
0.057547338 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 3 |
Hb_000291_180 |
0.0630556668 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 4 |
Hb_000320_190 |
0.0664386221 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 5 |
Hb_156850_100 |
0.0717208487 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 6 |
Hb_002107_050 |
0.0754739064 |
- |
- |
PREDICTED: prostaglandin E synthase 2 [Jatropha curcas] |
| 7 |
Hb_000108_020 |
0.077087745 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 8 |
Hb_003883_010 |
0.077914254 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 9 |
Hb_001882_010 |
0.0792984798 |
- |
- |
- |
| 10 |
Hb_012779_080 |
0.0797120484 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 11 |
Hb_010620_050 |
0.0800967422 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 12 |
Hb_002631_240 |
0.0828631164 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 13 |
Hb_000645_180 |
0.0848646057 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 14 |
Hb_000680_010 |
0.0853234716 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 15 |
Hb_002218_020 |
0.0865459006 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 16 |
Hb_068079_010 |
0.0881487844 |
- |
- |
- |
| 17 |
Hb_005062_060 |
0.0883905156 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 18 |
Hb_174865_040 |
0.0891003027 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_021596_020 |
0.0895312165 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 20 |
Hb_012438_030 |
0.0929487234 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |