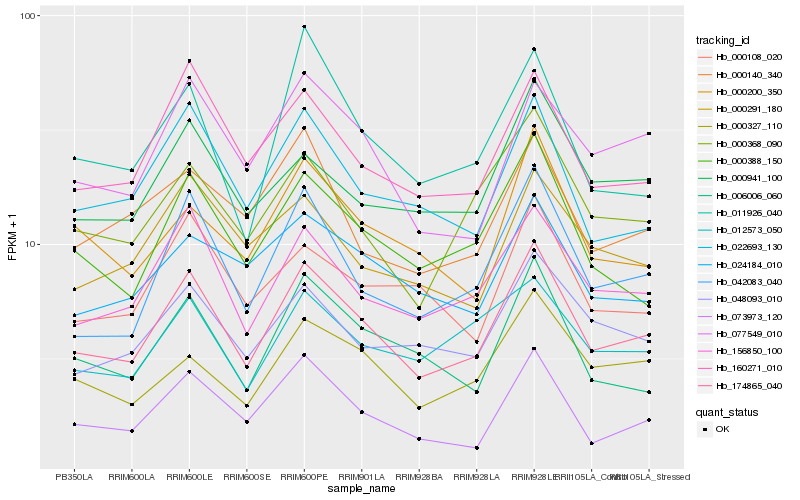
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174865_040 |
0.0 |
- |
- |
PREDICTED: crt homolog 1 isoform X2 [Jatropha curcas] |
| 2 |
Hb_156850_100 |
0.0797215712 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 3 |
Hb_042083_040 |
0.0802423611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_160271_010 |
0.0891003027 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_022693_130 |
0.0909195733 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_073973_120 |
0.091448851 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 7 |
Hb_000388_150 |
0.0920813581 |
- |
- |
hypothetical protein JCGZ_21651 [Jatropha curcas] |
| 8 |
Hb_077549_010 |
0.0957789013 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000327_110 |
0.0973530266 |
- |
- |
hypothetical protein JCGZ_06657 [Jatropha curcas] |
| 10 |
Hb_048093_010 |
0.0978102157 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_006006_060 |
0.0983909504 |
- |
- |
PREDICTED: triacylglycerol lipase 1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000368_090 |
0.1003956756 |
- |
- |
TPA: hypothetical protein ZEAMMB73_442906 [Zea mays] |
| 13 |
Hb_000941_100 |
0.1042738428 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 14 |
Hb_000200_350 |
0.1051492524 |
- |
- |
PREDICTED: uncharacterized protein LOC105636929 [Jatropha curcas] |
| 15 |
Hb_024184_010 |
0.1063491061 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 16 |
Hb_000108_020 |
0.1065743833 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 17 |
Hb_012573_050 |
0.1075344046 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 18 |
Hb_000291_180 |
0.108285283 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 19 |
Hb_000140_340 |
0.1087159803 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 20 |
Hb_011926_040 |
0.1087939103 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |