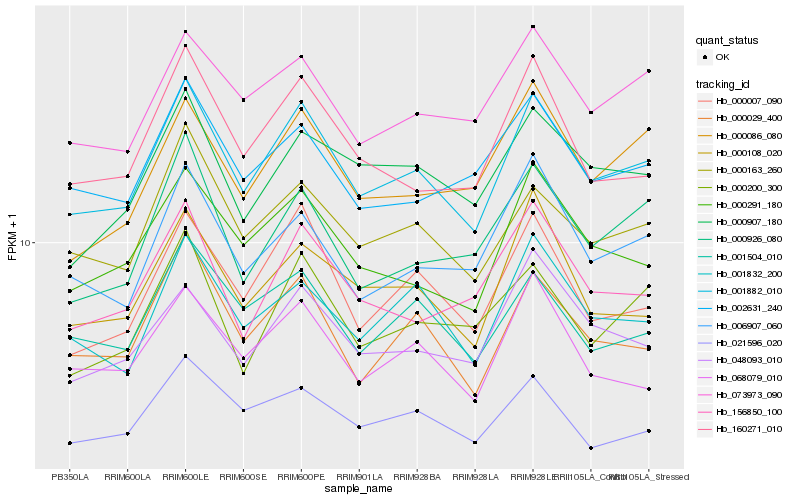
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001882_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_000163_260 |
0.0718985689 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 3 |
Hb_000291_180 |
0.0730021814 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 4 |
Hb_000029_400 |
0.0782478948 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000007_090 |
0.0786749059 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 6 |
Hb_160271_010 |
0.0792984798 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_073973_090 |
0.0798744412 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 8 |
Hb_001504_010 |
0.0807548771 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 9 |
Hb_001832_200 |
0.0810015027 |
- |
- |
PREDICTED: PCI domain-containing protein 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_002631_240 |
0.0831462544 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 11 |
Hb_006907_060 |
0.0839554452 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000108_020 |
0.0842834575 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 13 |
Hb_000907_180 |
0.0848460802 |
- |
- |
PREDICTED: uncharacterized protein LOC105641369 isoform X1 [Jatropha curcas] |
| 14 |
Hb_156850_100 |
0.0854169947 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 15 |
Hb_048093_010 |
0.0875632985 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_068079_010 |
0.0878751223 |
- |
- |
- |
| 17 |
Hb_000926_080 |
0.0886554278 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 18 |
Hb_000086_080 |
0.0888746583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_021596_020 |
0.089977438 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 20 |
Hb_000200_300 |
0.0912034203 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |