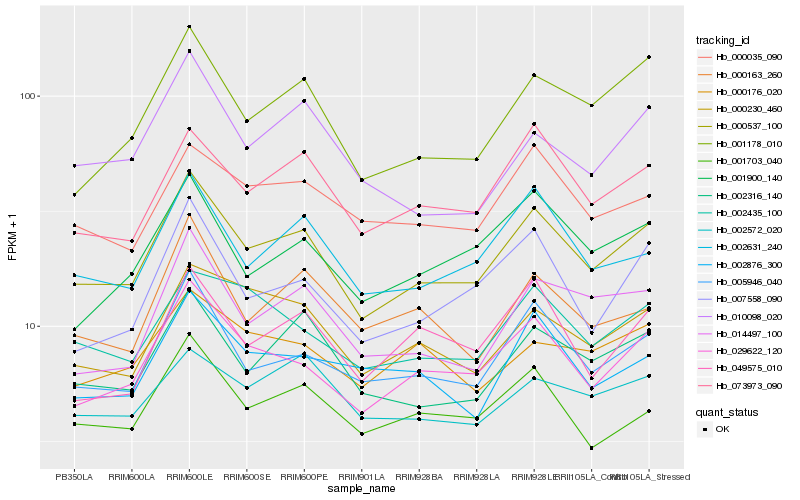
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000537_100 |
0.0 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_029622_120 |
0.0586205951 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2 [Jatropha curcas] |
| 3 |
Hb_005946_040 |
0.0642237135 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 4 |
Hb_073973_090 |
0.0688502064 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 5 |
Hb_002631_240 |
0.0721838551 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 6 |
Hb_002316_140 |
0.0724742811 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 7 |
Hb_001178_010 |
0.0758245837 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 8 |
Hb_001703_040 |
0.0788031264 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X2 [Jatropha curcas] |
| 9 |
Hb_014497_100 |
0.0795499947 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007558_090 |
0.079812741 |
- |
- |
PREDICTED: serine/threonine-protein kinase STN8, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_000230_460 |
0.0826793239 |
- |
- |
Speckle-type POZ protein, putative [Ricinus communis] |
| 12 |
Hb_000035_090 |
0.0828864144 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 13 |
Hb_002435_100 |
0.0833234636 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RNF217 [Jatropha curcas] |
| 14 |
Hb_001900_140 |
0.0841884552 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 15 |
Hb_002572_020 |
0.0892110627 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 16 |
Hb_049575_010 |
0.0902973159 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 17 |
Hb_000163_260 |
0.0906611293 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |
| 18 |
Hb_002876_300 |
0.0924548426 |
- |
- |
PREDICTED: histidinol-phosphate aminotransferase, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_010098_020 |
0.0925908273 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 20 |
Hb_000176_020 |
0.0936794821 |
- |
- |
PREDICTED: nuclear pore complex protein NUP43 [Jatropha curcas] |