| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
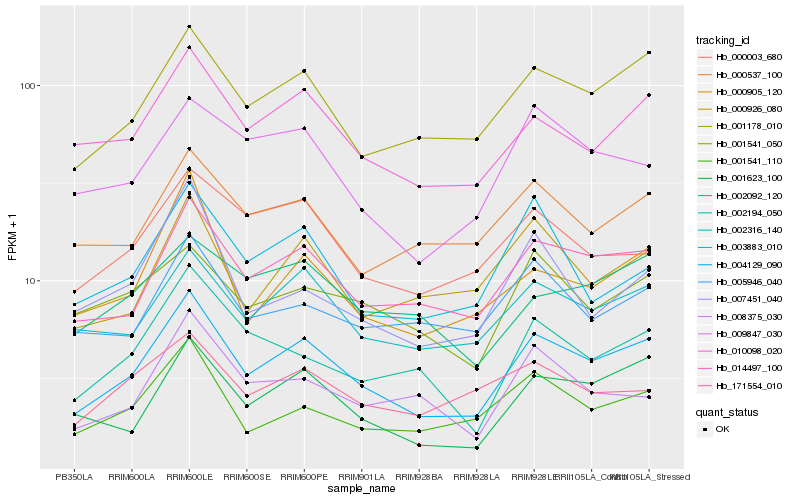
Hb_004129_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 2 |
Hb_014497_100 |
0.083956036 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 3 |
Hb_001178_010 |
0.0864187438 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 4 |
Hb_010098_020 |
0.1018609367 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_002316_140 |
0.1029100749 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 6 |
Hb_001623_100 |
0.1040803846 |
- |
- |
Hydroxyethylthiazole kinase, putative [Ricinus communis] |
| 7 |
Hb_001541_110 |
0.1119622735 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000905_120 |
0.112338806 |
- |
- |
PREDICTED: uncharacterized protein LOC105628677 [Jatropha curcas] |
| 9 |
Hb_000926_080 |
0.1166515082 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 10 |
Hb_002194_050 |
0.1225189902 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 11 |
Hb_009847_030 |
0.1227691717 |
- |
- |
zinc finger family protein [Populus trichocarpa] |
| 12 |
Hb_000537_100 |
0.123158688 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_003883_010 |
0.1244298479 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 14 |
Hb_000003_680 |
0.1247369476 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein H [Jatropha curcas] |
| 15 |
Hb_005946_040 |
0.1262518746 |
- |
- |
PREDICTED: probable protein phosphatase 2C 22 isoform X2 [Jatropha curcas] |
| 16 |
Hb_007451_040 |
0.1285221886 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 17 |
Hb_001541_050 |
0.1285486524 |
- |
- |
PREDICTED: serine/threonine-protein kinase AFC2-like [Jatropha curcas] |
| 18 |
Hb_171554_010 |
0.1291826354 |
- |
- |
PREDICTED: uncharacterized protein LOC105634071 [Jatropha curcas] |
| 19 |
Hb_002092_120 |
0.1295108922 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_008375_030 |
0.1298766659 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |