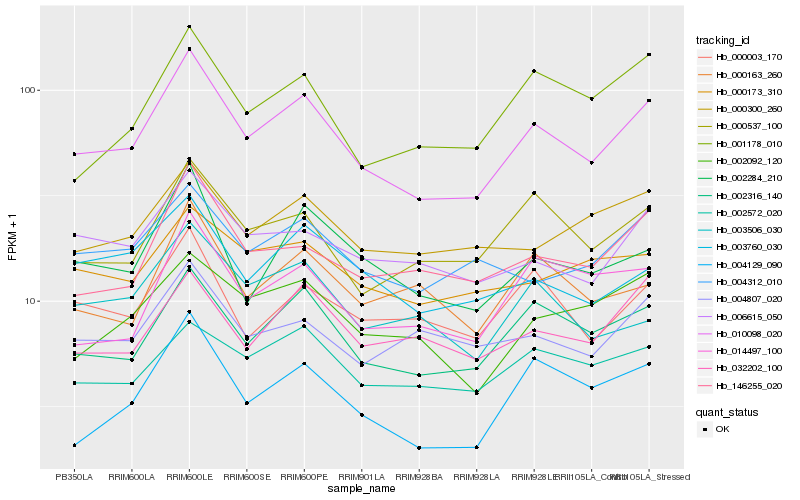
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010098_020 |
0.0 |
- |
- |
PREDICTED: protein BOLA4, chloroplastic/mitochondrial [Jatropha curcas] |
| 2 |
Hb_002316_140 |
0.0706003225 |
- |
- |
PREDICTED: rac-like GTP-binding protein 3 [Populus euphratica] |
| 3 |
Hb_000537_100 |
0.0925908273 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_002284_210 |
0.0938648551 |
- |
- |
PREDICTED: protein canopy-1 [Jatropha curcas] |
| 5 |
Hb_146255_020 |
0.0963941606 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 6 |
Hb_006615_050 |
0.0966752328 |
- |
- |
PREDICTED: uncharacterized protein LOC105640686 [Jatropha curcas] |
| 7 |
Hb_002572_020 |
0.099166085 |
transcription factor |
TF Family: SBP |
hypothetical protein POPTR_0005s28010g [Populus trichocarpa] |
| 8 |
Hb_003760_030 |
0.0992959802 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 9 |
Hb_000003_170 |
0.0994640118 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 10 |
Hb_002092_120 |
0.0995430879 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001178_010 |
0.1003484495 |
- |
- |
PREDICTED: uncharacterized protein LOC105629461 [Jatropha curcas] |
| 12 |
Hb_004312_010 |
0.1013923299 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X2 [Jatropha curcas] |
| 13 |
Hb_004129_090 |
0.1018609367 |
- |
- |
PREDICTED: uncharacterized protein LOC105646039 [Jatropha curcas] |
| 14 |
Hb_032202_100 |
0.1023401202 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 15 |
Hb_000300_260 |
0.1030011456 |
- |
- |
PREDICTED: replication factor C subunit 3 [Jatropha curcas] |
| 16 |
Hb_014497_100 |
0.1039844354 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000173_310 |
0.10504952 |
- |
- |
PREDICTED: uncharacterized protein LOC105631893 isoform X1 [Jatropha curcas] |
| 18 |
Hb_004807_020 |
0.1060272193 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 14b-like [Jatropha curcas] |
| 19 |
Hb_003506_030 |
0.1063359334 |
- |
- |
PREDICTED: ATP-dependent Clp protease ATP-binding subunit clpX-like, mitochondrial isoform X2 [Jatropha curcas] |
| 20 |
Hb_000163_260 |
0.106592311 |
- |
- |
PREDICTED: uncharacterized protein LOC105642518 [Jatropha curcas] |