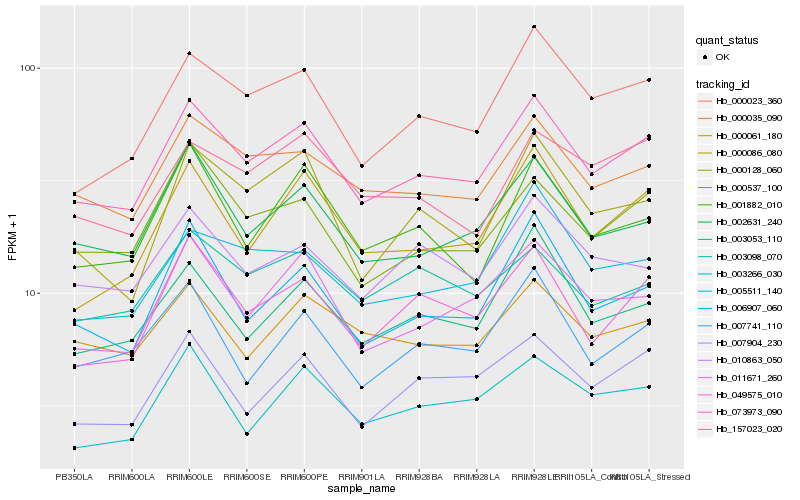
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_073973_090 |
0.0 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 2 |
Hb_000035_090 |
0.0579488889 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 3 |
Hb_000023_360 |
0.0623847873 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002631_240 |
0.063867513 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 5 |
Hb_011671_260 |
0.0645107309 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_049575_010 |
0.0659240126 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 7 |
Hb_000537_100 |
0.0688502064 |
- |
- |
Coiled-coil domain-containing protein, putative [Ricinus communis] |
| 8 |
Hb_006907_060 |
0.071183768 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_003053_110 |
0.0712107494 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000086_080 |
0.0714341963 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_007904_230 |
0.0762517987 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000061_180 |
0.0763506664 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 13 |
Hb_010863_050 |
0.0777626475 |
- |
- |
OTU domain-containing protein 6B, putative [Ricinus communis] |
| 14 |
Hb_000128_060 |
0.0788918707 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 15 |
Hb_005511_140 |
0.0794449227 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 16 |
Hb_001882_010 |
0.0798744412 |
- |
- |
- |
| 17 |
Hb_003098_070 |
0.0803635851 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 18 |
Hb_157023_020 |
0.0807201981 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_007741_110 |
0.0811209392 |
- |
- |
- |
| 20 |
Hb_003266_030 |
0.0813337615 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |