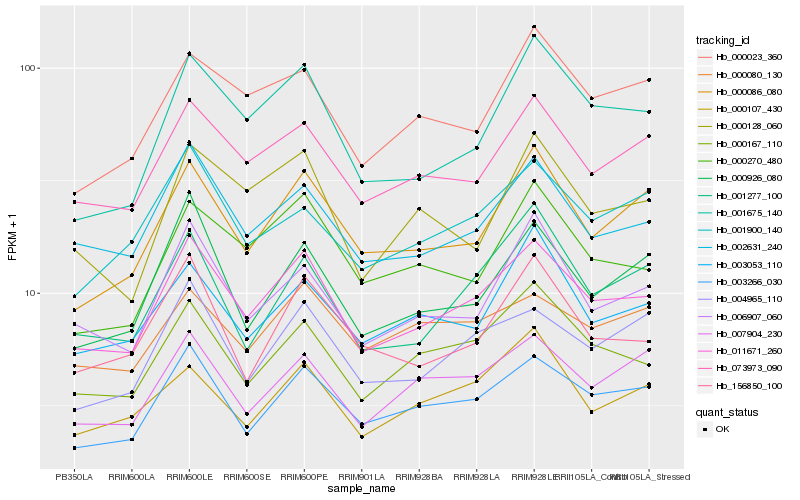
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011671_260 |
0.0 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003266_030 |
0.0601472589 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 3 |
Hb_002631_240 |
0.0618764207 |
- |
- |
JHL17M24.3 [Jatropha curcas] |
| 4 |
Hb_073973_090 |
0.0645107309 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 5 |
Hb_000167_110 |
0.0675589732 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_006907_060 |
0.0762113162 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_004965_110 |
0.0767036324 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 8 |
Hb_000086_080 |
0.0767476049 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007904_230 |
0.0812807225 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_001675_140 |
0.0818283846 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 11 |
Hb_000926_080 |
0.0835386371 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 12 |
Hb_000270_480 |
0.0836741208 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 13 |
Hb_001900_140 |
0.0861524873 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000023_360 |
0.086218667 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_156850_100 |
0.0868769751 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 16 |
Hb_000080_130 |
0.0890708729 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 17 |
Hb_003053_110 |
0.0892302416 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_000107_430 |
0.090275366 |
- |
- |
PREDICTED: brefeldin A-inhibited guanine nucleotide-exchange protein 1 [Jatropha curcas] |
| 19 |
Hb_000128_060 |
0.0906096007 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 20 |
Hb_001277_100 |
0.0907457898 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |