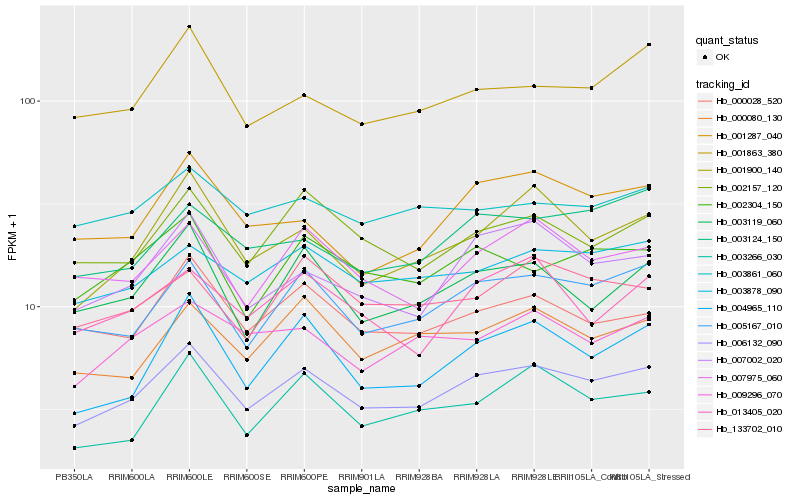
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006132_090 |
0.0 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 2 |
Hb_005167_010 |
0.0604952886 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001900_140 |
0.0717889344 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003878_090 |
0.0725699061 |
- |
- |
PREDICTED: polynucleotide 3'-phosphatase ZDP [Jatropha curcas] |
| 5 |
Hb_002157_120 |
0.0728053101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_007002_020 |
0.0745224427 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 7 |
Hb_003119_060 |
0.0755853903 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000028_520 |
0.0762631417 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 9 |
Hb_007975_060 |
0.0769690574 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 10 |
Hb_004965_110 |
0.0775727176 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 11 |
Hb_000080_130 |
0.0779388875 |
- |
- |
PREDICTED: TBC1 domain family member 22B [Jatropha curcas] |
| 12 |
Hb_002304_150 |
0.078248694 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001863_380 |
0.0796987755 |
- |
- |
PREDICTED: 14-3-3 protein 7 [Jatropha curcas] |
| 14 |
Hb_009296_070 |
0.0799850936 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001287_040 |
0.0822592627 |
- |
- |
PREDICTED: kinesin-like protein NACK1 [Jatropha curcas] |
| 16 |
Hb_003266_030 |
0.0838834543 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 17 |
Hb_003124_150 |
0.0844289728 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 28-like [Jatropha curcas] |
| 18 |
Hb_133702_010 |
0.0858721636 |
- |
- |
PREDICTED: reticulon-4-interacting protein 1, mitochondrial isoform X1 [Jatropha curcas] |
| 19 |
Hb_013405_020 |
0.085963765 |
- |
- |
- |
| 20 |
Hb_003861_060 |
0.0867880629 |
- |
- |
PREDICTED: treacle protein [Jatropha curcas] |