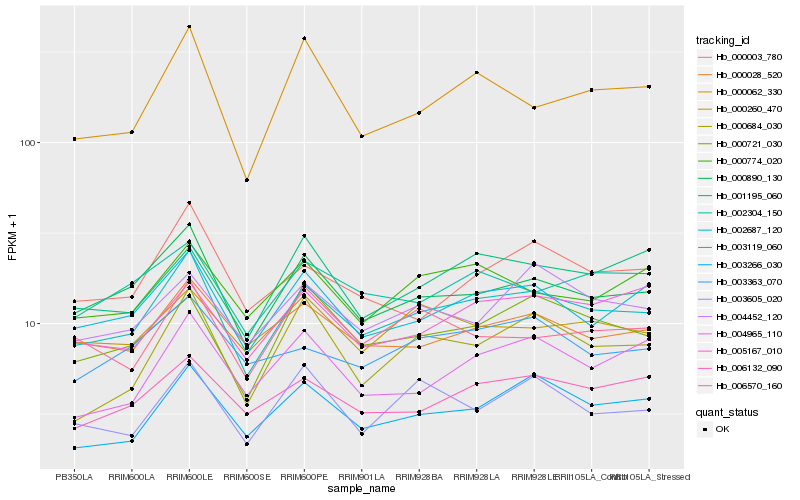
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002687_120 |
0.0 |
- |
- |
PREDICTED: lipoyl synthase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000890_130 |
0.0643169721 |
- |
- |
PREDICTED: probable serine/threonine-protein phosphatase 2A regulatory subunit B'' subunit TON2 [Phoenix dactylifera] |
| 3 |
Hb_003119_060 |
0.0728973356 |
- |
- |
PREDICTED: dicarboxylate transporter 2.1, chloroplastic [Jatropha curcas] |
| 4 |
Hb_003266_030 |
0.0836049243 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 5 |
Hb_000028_520 |
0.0870715738 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 6 |
Hb_000721_030 |
0.0892315492 |
rubber biosynthesis |
Gene Name: Mevalonate diphosphate decarboxylase |
mevalonate diphosphate decarboxylase [Hevea brasiliensis] |
| 7 |
Hb_006132_090 |
0.0894645598 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000062_330 |
0.0925470551 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_002304_150 |
0.0936517005 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_000003_780 |
0.0942332532 |
- |
- |
hexokinase [Manihot esculenta] |
| 11 |
Hb_000774_020 |
0.0963590668 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 12 |
Hb_003363_070 |
0.0975050256 |
- |
- |
- |
| 13 |
Hb_005167_010 |
0.0997828756 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000684_030 |
0.1003123532 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 15 |
Hb_003605_020 |
0.1012252625 |
- |
- |
exocyst complex component sec6, putative [Ricinus communis] |
| 16 |
Hb_004452_120 |
0.1012773464 |
- |
- |
PREDICTED: uncharacterized protein LOC105639574 [Jatropha curcas] |
| 17 |
Hb_000260_470 |
0.1014056447 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 18 |
Hb_004965_110 |
0.1021085579 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 19 |
Hb_006570_160 |
0.1022769628 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 20 |
Hb_001195_060 |
0.1025881372 |
- |
- |
Protein yrdA, putative [Ricinus communis] |