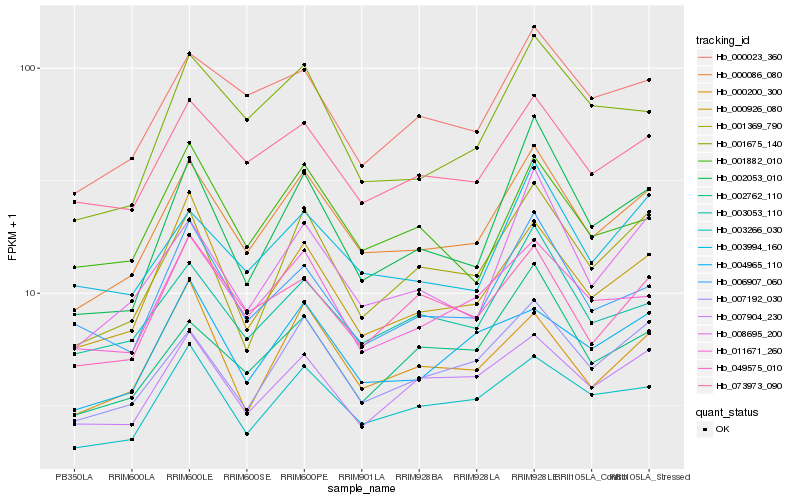
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_007192_030 |
0.0690798603 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 3 |
Hb_073973_090 |
0.0714341963 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 4 |
Hb_003266_030 |
0.0720479947 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 5 |
Hb_011671_260 |
0.0767476049 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000023_360 |
0.0769049692 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003053_110 |
0.077121648 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_008695_200 |
0.0779267345 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 9 |
Hb_000926_080 |
0.0784359197 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 10 |
Hb_001369_790 |
0.0816485964 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003994_160 |
0.0821768327 |
- |
- |
PREDICTED: uncharacterized protein LOC105131691 isoform X5 [Populus euphratica] |
| 12 |
Hb_001675_140 |
0.0842616682 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 13 |
Hb_007904_230 |
0.085598146 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 14 |
Hb_006907_060 |
0.0867654663 |
- |
- |
Alternative oxidase 4, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_049575_010 |
0.0867741066 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 16 |
Hb_004965_110 |
0.086822447 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 17 |
Hb_002053_010 |
0.0871032717 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_002762_110 |
0.0887151687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001882_010 |
0.0888746583 |
- |
- |
- |
| 20 |
Hb_000200_300 |
0.0894998419 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |