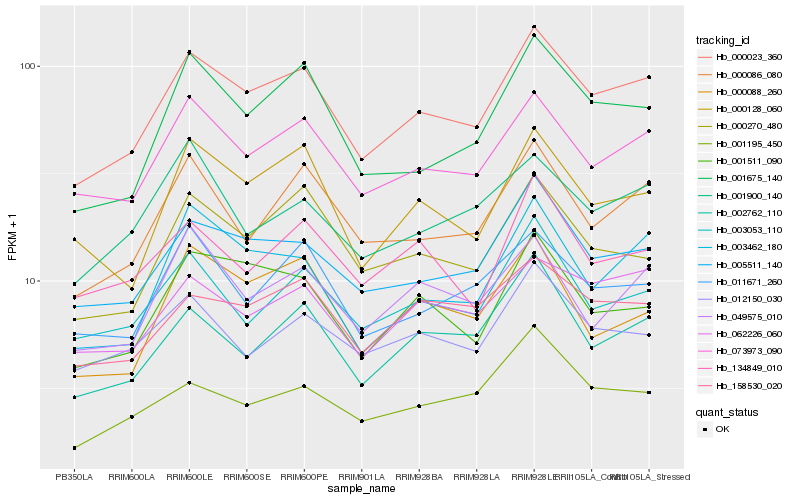
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000023_360 |
0.0 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_073973_090 |
0.0623847873 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 3 |
Hb_005511_140 |
0.0646167777 |
- |
- |
Ubiquinone biosynthesis protein coq-8, putative [Ricinus communis] |
| 4 |
Hb_000086_080 |
0.0769049692 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001675_140 |
0.0785322591 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 6 |
Hb_062226_060 |
0.0795374363 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein B-like [Jatropha curcas] |
| 7 |
Hb_158530_020 |
0.0812843687 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 8 |
Hb_000128_060 |
0.0813119476 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 9 |
Hb_001511_090 |
0.0813452279 |
- |
- |
PREDICTED: uncharacterized protein LOC105645861 [Jatropha curcas] |
| 10 |
Hb_002762_110 |
0.0817201681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003053_110 |
0.0818304694 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000270_480 |
0.0821345962 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 13 |
Hb_003462_180 |
0.0860724997 |
transcription factor |
TF Family: GNAT |
PREDICTED: probable acetyltransferase NATA1-like [Jatropha curcas] |
| 14 |
Hb_011671_260 |
0.086218667 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_012150_030 |
0.0895404274 |
- |
- |
PREDICTED: aspartate--tRNA ligase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_049575_010 |
0.0903893627 |
- |
- |
hypothetical protein POPTR_0001s15330g [Populus trichocarpa] |
| 17 |
Hb_001195_450 |
0.0917205145 |
- |
- |
PREDICTED: uncharacterized protein LOC105633771 [Jatropha curcas] |
| 18 |
Hb_000088_260 |
0.0934848549 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 19 |
Hb_134849_010 |
0.0937590936 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 20 |
Hb_001900_140 |
0.0945660436 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |