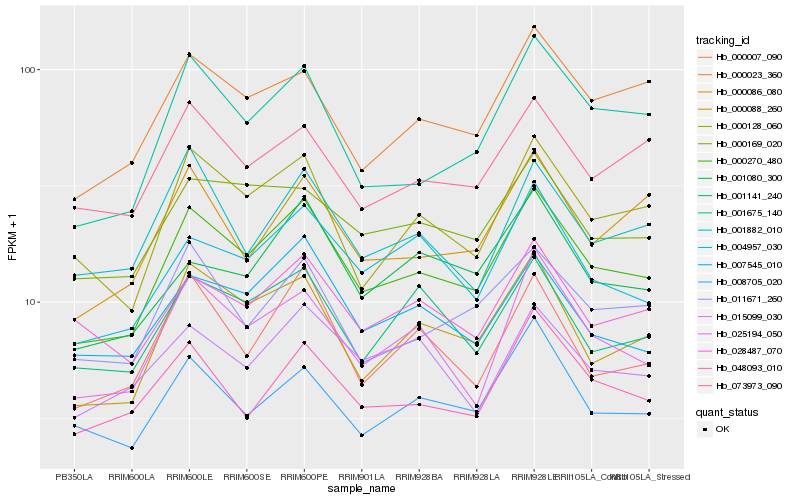
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000270_480 |
0.0 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 2 |
Hb_000088_260 |
0.0685866091 |
- |
- |
PREDICTED: uncharacterized protein LOC105636987 [Jatropha curcas] |
| 3 |
Hb_000128_060 |
0.0737737604 |
- |
- |
PREDICTED: sodium/pyruvate cotransporter BASS2, chloroplastic isoform X1 [Vitis vinifera] |
| 4 |
Hb_025194_050 |
0.0772301078 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001675_140 |
0.0794263109 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 6 |
Hb_007545_010 |
0.0818801281 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 7 |
Hb_000023_360 |
0.0821345962 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004957_030 |
0.0821362309 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 9 |
Hb_000169_020 |
0.0830917001 |
transcription factor |
TF Family: C2C2-CO-like |
hypothetical protein RCOM_0555710 [Ricinus communis] |
| 10 |
Hb_048093_010 |
0.0833629833 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 11 |
Hb_011671_260 |
0.0836741208 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000007_090 |
0.0849121045 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 13 |
Hb_008705_020 |
0.0877890083 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_001141_240 |
0.088737044 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_073973_090 |
0.0889353258 |
- |
- |
Aminotransferase ybdL, putative [Ricinus communis] |
| 16 |
Hb_028487_070 |
0.0903996761 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001080_300 |
0.0905095634 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 18 |
Hb_000086_080 |
0.0905480619 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001882_010 |
0.0914331 |
- |
- |
- |
| 20 |
Hb_015099_030 |
0.0931480924 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |