| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
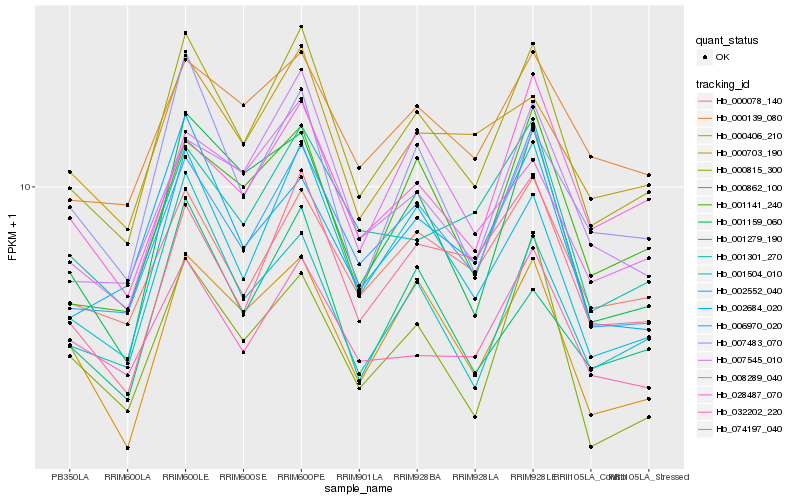
Hb_000815_300 |
0.0 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 2 |
Hb_002552_040 |
0.0501906924 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 3 |
Hb_006970_020 |
0.0528012064 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 4 |
Hb_000406_210 |
0.0615236989 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 5 |
Hb_000078_140 |
0.0653859755 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 6 |
Hb_001141_240 |
0.0656919492 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_074197_040 |
0.0676424344 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 8 |
Hb_001504_010 |
0.0683886798 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 9 |
Hb_001159_060 |
0.0688144153 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 10 |
Hb_000862_100 |
0.0693362611 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_007545_010 |
0.0695634603 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 12 |
Hb_000703_190 |
0.0710517308 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001301_270 |
0.0718799173 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 14 |
Hb_001279_190 |
0.072600586 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 15 |
Hb_007483_070 |
0.0726406641 |
- |
- |
PREDICTED: intersectin-1 isoform X1 [Populus euphratica] |
| 16 |
Hb_002684_020 |
0.0730575603 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 17 |
Hb_000139_080 |
0.0736487823 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_032202_220 |
0.0750468248 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_008289_040 |
0.0752812612 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 20 |
Hb_028487_070 |
0.075343036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |