| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
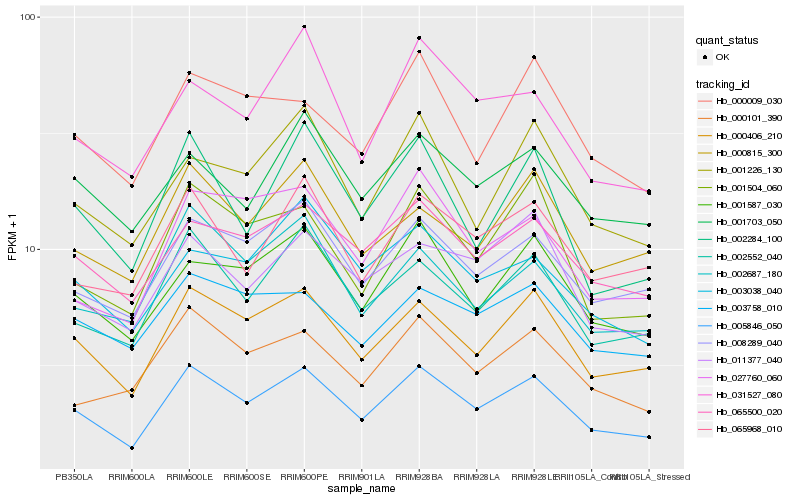
Hb_005846_050 |
0.0 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 2 |
Hb_000406_210 |
0.0554307942 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 3 |
Hb_002552_040 |
0.0742500742 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 4 |
Hb_001587_030 |
0.0760830383 |
- |
- |
hypothetical protein JCGZ_09892 [Jatropha curcas] |
| 5 |
Hb_008289_040 |
0.0770102286 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 6 |
Hb_065500_020 |
0.0806830649 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 7 |
Hb_001504_060 |
0.0828446744 |
- |
- |
PREDICTED: nuclear pore complex protein NUP93A-like [Jatropha curcas] |
| 8 |
Hb_065968_010 |
0.0883251863 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 9 |
Hb_000101_390 |
0.0885686693 |
- |
- |
PREDICTED: mucin-5B [Jatropha curcas] |
| 10 |
Hb_002284_100 |
0.0893234386 |
- |
- |
ribophorin, putative [Ricinus communis] |
| 11 |
Hb_011377_040 |
0.0896128402 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 12 |
Hb_001703_050 |
0.091762684 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 13 |
Hb_001226_130 |
0.0919138222 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 14 |
Hb_000815_300 |
0.092103661 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 15 |
Hb_027760_060 |
0.0927586696 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375-like [Jatropha curcas] |
| 16 |
Hb_000009_030 |
0.0931609876 |
- |
- |
PREDICTED: splicing factor U2af large subunit B isoform X1 [Jatropha curcas] |
| 17 |
Hb_002687_180 |
0.0938240767 |
- |
- |
PREDICTED: phytochrome-associated serine/threonine-protein phosphatase [Jatropha curcas] |
| 18 |
Hb_031527_080 |
0.0964029215 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 19 |
Hb_003038_040 |
0.096638341 |
transcription factor |
TF Family: TUB |
phosphoric diester hydrolase, putative [Ricinus communis] |
| 20 |
Hb_003758_010 |
0.0971766736 |
transcription factor |
TF Family: VOZ |
PREDICTED: transcription factor VOZ1 isoform X1 [Jatropha curcas] |