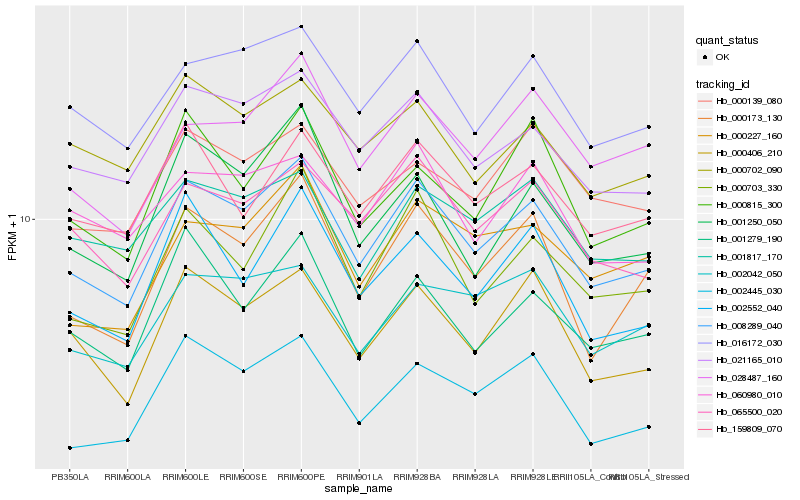
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008289_040 |
0.0 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 2 |
Hb_021165_010 |
0.0539433119 |
- |
- |
PREDICTED: splicing factor U2af large subunit B-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_016172_030 |
0.0601858779 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SEC [Jatropha curcas] |
| 4 |
Hb_000173_130 |
0.0620228151 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 5 |
Hb_065500_020 |
0.0631820119 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 6 |
Hb_000406_210 |
0.0645796606 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 7 |
Hb_000703_330 |
0.0660388843 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_028487_160 |
0.0702204397 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 9 |
Hb_002552_040 |
0.071025179 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 10 |
Hb_001250_050 |
0.0714108714 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 11 |
Hb_159809_070 |
0.0716526723 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 12 |
Hb_000227_160 |
0.072038278 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 13 |
Hb_002042_050 |
0.0731684937 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002445_030 |
0.0747094404 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 15 |
Hb_000702_090 |
0.0750769457 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 16 |
Hb_001817_170 |
0.07522773 |
- |
- |
PREDICTED: AP-1 complex subunit gamma-2-like [Jatropha curcas] |
| 17 |
Hb_000815_300 |
0.0752812612 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 18 |
Hb_000139_080 |
0.0752974068 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001279_190 |
0.0753376736 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 20 |
Hb_060980_010 |
0.0753901743 |
- |
- |
PREDICTED: BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1-like isoform X1 [Populus euphratica] |