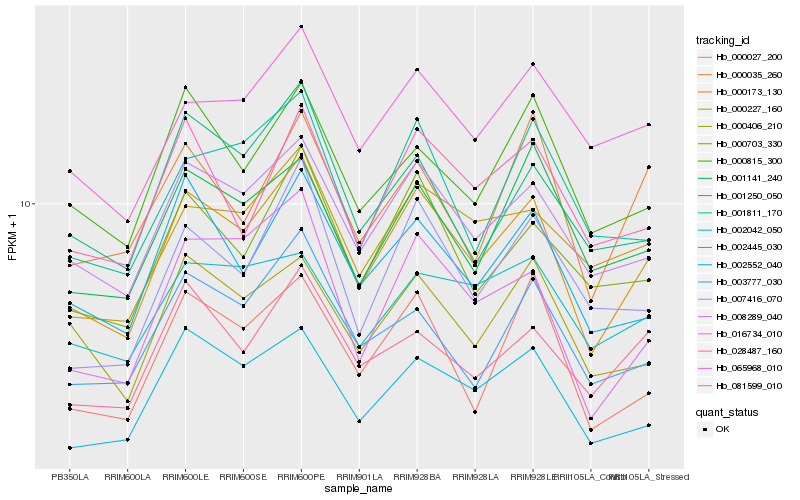
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000173_130 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 2 |
Hb_008289_040 |
0.0620228151 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 3 |
Hb_002445_030 |
0.0723441493 |
- |
- |
hypothetical protein JCGZ_12656 [Jatropha curcas] |
| 4 |
Hb_002552_040 |
0.0801505474 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 5 |
Hb_000703_330 |
0.0828212323 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000815_300 |
0.0861370015 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 7 |
Hb_016734_010 |
0.0863995962 |
- |
- |
PREDICTED: probable DNA-3-methyladenine glycosylase 2 [Jatropha curcas] |
| 8 |
Hb_000406_210 |
0.0866979414 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 9 |
Hb_001141_240 |
0.0867345249 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_065968_010 |
0.0877866461 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000227_160 |
0.08853699 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A [Jatropha curcas] |
| 12 |
Hb_081599_010 |
0.0894755638 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_003777_030 |
0.0896323514 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 14 |
Hb_028487_160 |
0.0899790712 |
- |
- |
clathrin assembly protein, putative [Ricinus communis] |
| 15 |
Hb_000027_200 |
0.0904120035 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 16 |
Hb_007416_070 |
0.0915528319 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 17 |
Hb_001811_170 |
0.093166382 |
- |
- |
dynamin, putative [Ricinus communis] |
| 18 |
Hb_001250_050 |
0.0942786927 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002042_050 |
0.0958218296 |
- |
- |
PREDICTED: uncharacterized protein LOC105647554 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000035_260 |
0.0970148936 |
- |
- |
PREDICTED: probable protein phosphatase 2C 60 [Jatropha curcas] |