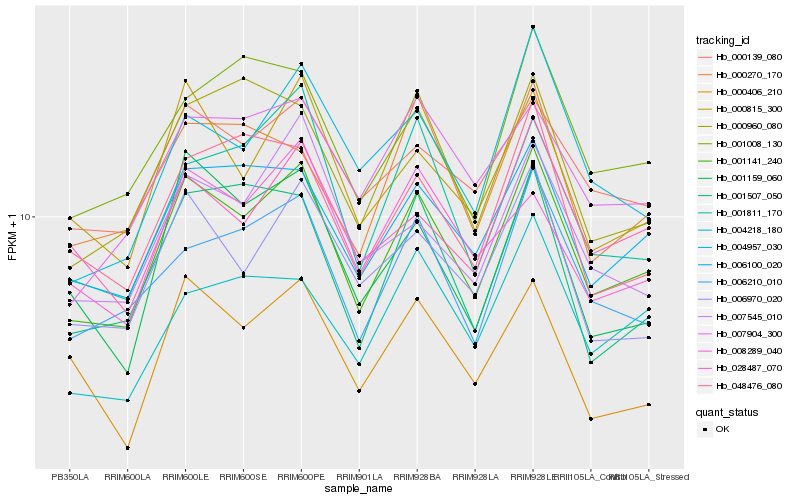
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_240 |
0.0 |
- |
- |
PREDICTED: folate-biopterin transporter 1, chloroplastic [Jatropha curcas] |
| 2 |
Hb_006100_020 |
0.0584803561 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 3 |
Hb_000815_300 |
0.0656919492 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 4 |
Hb_006210_010 |
0.0670720401 |
- |
- |
PREDICTED: probable acyl-activating enzyme 17, peroxisomal isoform X2 [Jatropha curcas] |
| 5 |
Hb_000139_080 |
0.0695123413 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001811_170 |
0.0716654843 |
- |
- |
dynamin, putative [Ricinus communis] |
| 7 |
Hb_006970_020 |
0.071979539 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 8 |
Hb_007545_010 |
0.0746444056 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 9 |
Hb_048476_080 |
0.0753405086 |
transcription factor |
TF Family: ARR-B |
two-component sensor histidine kinase bacteria, putative [Ricinus communis] |
| 10 |
Hb_001008_130 |
0.0756279189 |
transcription factor |
TF Family: mTERF |
PREDICTED: cytochrome c1-2, heme protein, mitochondrial-like [Elaeis guineensis] |
| 11 |
Hb_028487_070 |
0.0760693446 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000960_080 |
0.0766380071 |
- |
- |
PREDICTED: topless-related protein 1-like isoform X2 [Jatropha curcas] |
| 13 |
Hb_001507_050 |
0.0767101057 |
- |
- |
PREDICTED: cysteine desulfurase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000406_210 |
0.0768768916 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 15 |
Hb_008289_040 |
0.0769130967 |
- |
- |
PREDICTED: suppressor of mec-8 and unc-52 protein homolog 1 [Jatropha curcas] |
| 16 |
Hb_004218_180 |
0.0772788562 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |
| 17 |
Hb_001159_060 |
0.0778245727 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 18 |
Hb_004957_030 |
0.0782737236 |
- |
- |
hydroxyproline-rich glycoprotein [Populus trichocarpa] |
| 19 |
Hb_007904_300 |
0.0782885013 |
- |
- |
copine, putative [Ricinus communis] |
| 20 |
Hb_000270_170 |
0.0784655917 |
- |
- |
PREDICTED: formin-like protein 5 [Jatropha curcas] |