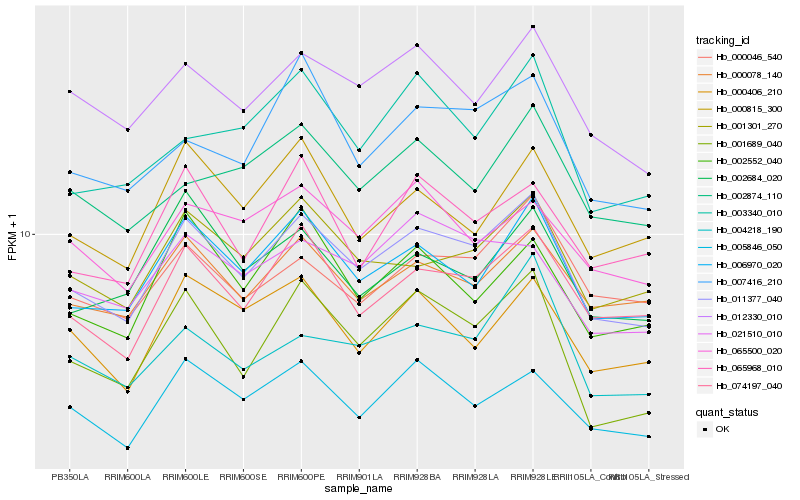
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011377_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647286 [Jatropha curcas] |
| 2 |
Hb_006970_020 |
0.0632750383 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 3 |
Hb_074197_040 |
0.0690463818 |
- |
- |
PREDICTED: SNARE-interacting protein KEULE [Jatropha curcas] |
| 4 |
Hb_012330_010 |
0.0747000746 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 12-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_001301_270 |
0.0768303315 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 6 |
Hb_000078_140 |
0.0790601626 |
- |
- |
PREDICTED: WD repeat-containing protein 11 [Jatropha curcas] |
| 7 |
Hb_000046_540 |
0.0869058498 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 34-like [Populus euphratica] |
| 8 |
Hb_001689_040 |
0.0885903734 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 1-like [Jatropha curcas] |
| 9 |
Hb_005846_050 |
0.0896128402 |
- |
- |
PREDICTED: calmodulin-binding transcription activator 3 [Jatropha curcas] |
| 10 |
Hb_007416_210 |
0.0921698398 |
- |
- |
PREDICTED: coatomer subunit beta-1 [Jatropha curcas] |
| 11 |
Hb_000406_210 |
0.0929262449 |
- |
- |
PREDICTED: protein HASTY 1 [Jatropha curcas] |
| 12 |
Hb_000815_300 |
0.0931572716 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 13 |
Hb_002684_020 |
0.0936080333 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 14 |
Hb_021510_010 |
0.0936498057 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 15 |
Hb_065500_020 |
0.0943528152 |
- |
- |
Exocyst complex component sec3A isoform 1 [Theobroma cacao] |
| 16 |
Hb_002874_110 |
0.094768483 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 17 |
Hb_002552_040 |
0.0950060176 |
- |
- |
PREDICTED: uncharacterized protein LOC105641220 [Jatropha curcas] |
| 18 |
Hb_065968_010 |
0.0952303838 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003340_010 |
0.095537842 |
- |
- |
PREDICTED: ubiquitin-activating enzyme E1 1 [Vitis vinifera] |
| 20 |
Hb_004218_190 |
0.0962215453 |
transcription factor |
TF Family: SNF2 |
PREDICTED: F-box protein At3g54460 isoform X1 [Jatropha curcas] |