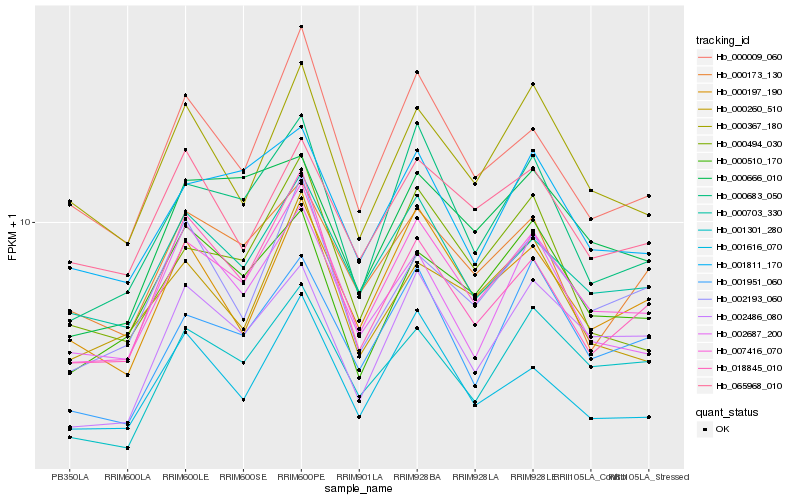
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_070 |
0.0 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 2 |
Hb_000703_330 |
0.06538856 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000683_050 |
0.0670818488 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 4 |
Hb_000009_060 |
0.0766370667 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 5 |
Hb_000197_190 |
0.0785551308 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000367_180 |
0.0821841544 |
- |
- |
Heparanase-2, putative [Ricinus communis] |
| 7 |
Hb_001811_170 |
0.0823319984 |
- |
- |
dynamin, putative [Ricinus communis] |
| 8 |
Hb_002486_080 |
0.0874286916 |
- |
- |
PREDICTED: cytosolic endo-beta-N-acetylglucosaminidase isoform X1 [Jatropha curcas] |
| 9 |
Hb_002193_060 |
0.0876388071 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 10 |
Hb_018845_010 |
0.0878104486 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 11 |
Hb_001616_070 |
0.0882389394 |
- |
- |
PREDICTED: uncharacterized protein LOC105644365 [Jatropha curcas] |
| 12 |
Hb_001301_280 |
0.0885374848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000510_170 |
0.0888672844 |
- |
- |
Glycosyltransferase QUASIMODO1, putative [Ricinus communis] |
| 14 |
Hb_000494_030 |
0.0891752207 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 15 |
Hb_001951_060 |
0.0912252639 |
- |
- |
PREDICTED: UDP-N-acetylglucosamine transferase subunit ALG14 [Jatropha curcas] |
| 16 |
Hb_002687_200 |
0.0915525758 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000173_130 |
0.0915528319 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase LUL4 isoform X1 [Jatropha curcas] |
| 18 |
Hb_065968_010 |
0.0917392603 |
- |
- |
PREDICTED: probable glycosyltransferase At3g07620 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000260_510 |
0.0925561894 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 20 |
Hb_000666_010 |
0.0935124155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |