| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
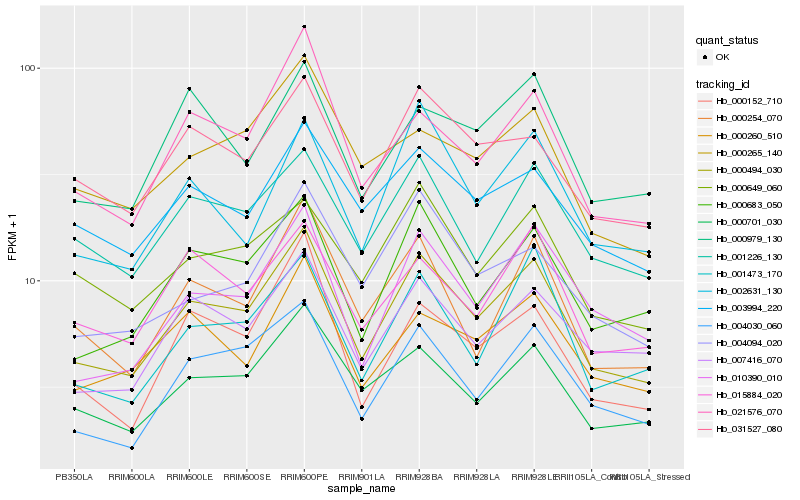
Hb_000494_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 2 |
Hb_000701_030 |
0.0779698188 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |
| 3 |
Hb_000683_050 |
0.0798980745 |
- |
- |
PREDICTED: probable carbohydrate esterase At4g34215 [Jatropha curcas] |
| 4 |
Hb_004030_060 |
0.0835456674 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 5 |
Hb_010390_010 |
0.0870410566 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 6 |
Hb_021576_070 |
0.0881243382 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 7 |
Hb_007416_070 |
0.0891752207 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_07038 [Jatropha curcas] |
| 8 |
Hb_001226_130 |
0.0906215558 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 9 |
Hb_000254_070 |
0.0912079159 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 10 |
Hb_000260_510 |
0.0931927793 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X3 [Jatropha curcas] |
| 11 |
Hb_031527_080 |
0.0940044317 |
- |
- |
utp-glucose-1-phosphate uridylyltransferase, putative [Ricinus communis] |
| 12 |
Hb_001473_170 |
0.0941895276 |
- |
- |
PREDICTED: non-lysosomal glucosylceramidase [Jatropha curcas] |
| 13 |
Hb_003994_220 |
0.0951378582 |
- |
- |
PREDICTED: V-type proton ATPase catalytic subunit A [Jatropha curcas] |
| 14 |
Hb_015884_020 |
0.0978132925 |
- |
- |
PREDICTED: protein PIR [Jatropha curcas] |
| 15 |
Hb_000649_060 |
0.0978756079 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 16 |
Hb_002631_130 |
0.0994030077 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 17 |
Hb_000979_130 |
0.103731038 |
- |
- |
PREDICTED: phosphoglucomutase, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_000152_710 |
0.1038107276 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 19 |
Hb_004094_020 |
0.1039611531 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000265_140 |
0.1043879791 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 12 [Jatropha curcas] |