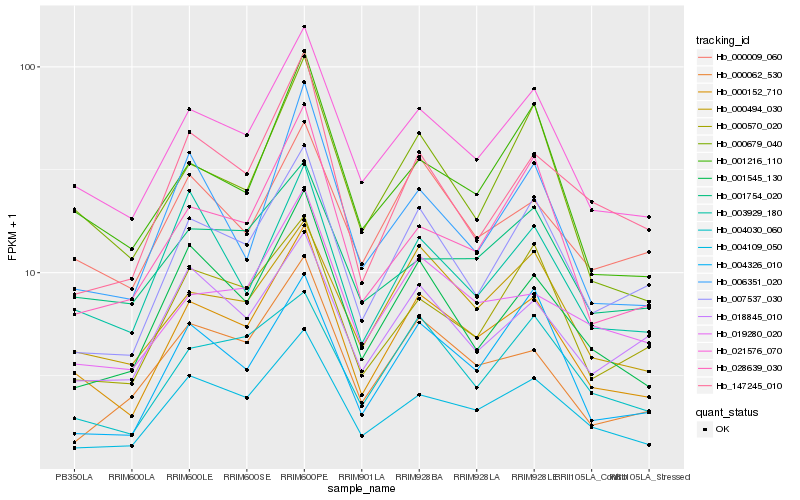
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_710 |
0.0 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 1 [Jatropha curcas] |
| 2 |
Hb_021576_070 |
0.0660960938 |
- |
- |
hypothetical protein PRUPE_ppa002736mg [Prunus persica] |
| 3 |
Hb_004109_050 |
0.0814188484 |
- |
- |
PREDICTED: putative callose synthase 8 [Jatropha curcas] |
| 4 |
Hb_007537_030 |
0.0966988198 |
- |
- |
PREDICTED: gamma aminobutyrate transaminase 3, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001545_130 |
0.1005915761 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 6 |
Hb_019280_020 |
0.1037445983 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000494_030 |
0.1038107276 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |
| 8 |
Hb_000679_040 |
0.1065790365 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 9 |
Hb_001216_110 |
0.1075226847 |
- |
- |
PREDICTED: protein GPR107-like [Jatropha curcas] |
| 10 |
Hb_000009_060 |
0.1120051305 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 1B [Jatropha curcas] |
| 11 |
Hb_028639_030 |
0.1141429074 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 2-like [Jatropha curcas] |
| 12 |
Hb_000062_530 |
0.1144591797 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 13 |
Hb_003929_180 |
0.1149287998 |
- |
- |
PREDICTED: probable sugar phosphate/phosphate translocator At3g17430 isoform X1 [Glycine max] |
| 14 |
Hb_006351_020 |
0.1154047552 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase S.4-like [Jatropha curcas] |
| 15 |
Hb_000570_020 |
0.1160463525 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX9H isoform X1 [Jatropha curcas] |
| 16 |
Hb_004030_060 |
0.1189394634 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 17 |
Hb_147245_010 |
0.1200291419 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 18 |
Hb_018845_010 |
0.1208201072 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 19 |
Hb_004326_010 |
0.1216661418 |
- |
- |
cationic amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_001754_020 |
0.1227033207 |
- |
- |
PREDICTED: kinesin-13A isoform X1 [Jatropha curcas] |