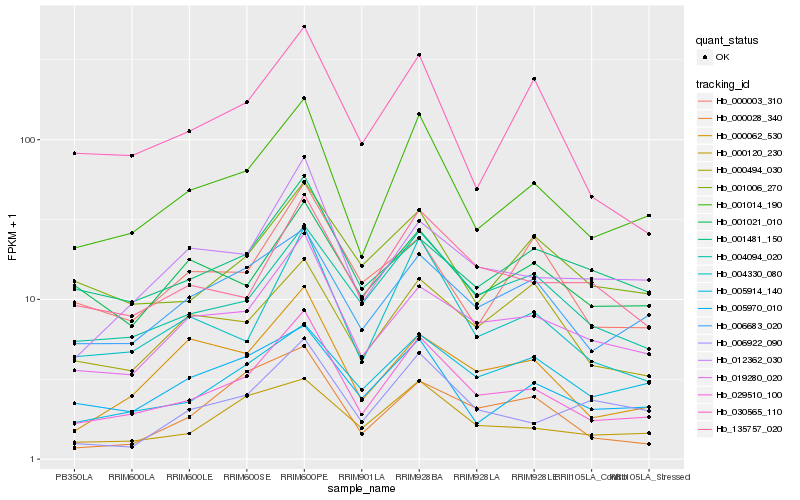
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029510_100 |
0.0 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 2 |
Hb_001014_190 |
0.0761033679 |
- |
- |
PREDICTED: uncharacterized protein LOC105647294 [Jatropha curcas] |
| 3 |
Hb_004330_080 |
0.1063321943 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_019280_020 |
0.1066097505 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK14 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004094_020 |
0.1214841766 |
- |
- |
Pyruvate kinase family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_000120_230 |
0.1220659419 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33990-like [Jatropha curcas] |
| 7 |
Hb_012362_030 |
0.1253592806 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Populus euphratica] |
| 8 |
Hb_006683_020 |
0.1275402401 |
- |
- |
PREDICTED: nucleosome assembly protein 1;4 [Jatropha curcas] |
| 9 |
Hb_135757_020 |
0.1291855609 |
- |
- |
hypothetical protein AMTR_s00420p00013340 [Amborella trichopoda] |
| 10 |
Hb_000062_530 |
0.1361342297 |
- |
- |
PREDICTED: GDP-mannose transporter GONST3-like [Populus euphratica] |
| 11 |
Hb_030565_110 |
0.1395450147 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 12 |
Hb_005970_010 |
0.1424484874 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001006_270 |
0.1438468472 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 14 |
Hb_005914_140 |
0.1476680988 |
- |
- |
PREDICTED: uncharacterized protein LOC105632183 [Jatropha curcas] |
| 15 |
Hb_001481_150 |
0.1483431641 |
- |
- |
hypothetical protein POPTR_0001s15440g [Populus trichocarpa] |
| 16 |
Hb_006922_090 |
0.1516753957 |
- |
- |
PREDICTED: interaptin [Jatropha curcas] |
| 17 |
Hb_000003_310 |
0.1529327123 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 18 |
Hb_000028_340 |
0.1538358085 |
- |
- |
PREDICTED: potassium channel SKOR-like [Prunus mume] |
| 19 |
Hb_001021_010 |
0.1538875211 |
- |
- |
PREDICTED: bifunctional protein FolD 2 [Jatropha curcas] |
| 20 |
Hb_000494_030 |
0.1542551383 |
- |
- |
PREDICTED: uncharacterized protein LOC105643196 [Jatropha curcas] |