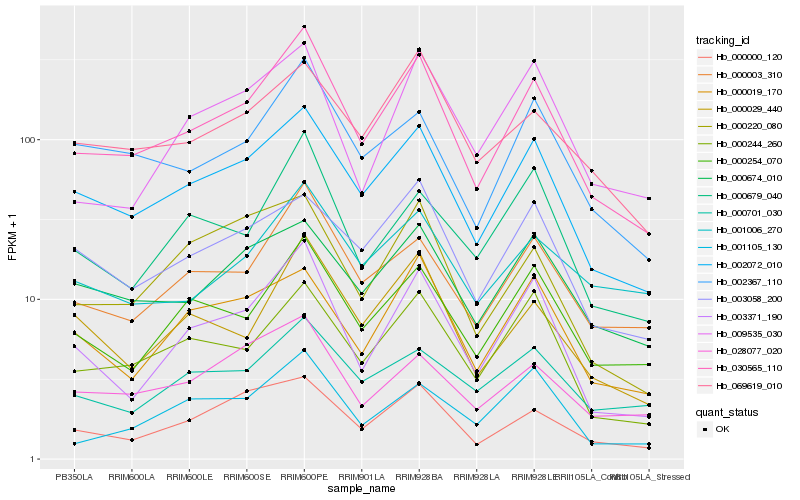
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030565_110 |
0.0 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 2 |
Hb_002072_010 |
0.0728092536 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003371_190 |
0.0952537718 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 4 |
Hb_000003_310 |
0.098832506 |
- |
- |
PREDICTED: glycosyltransferase-like KOBITO 1 [Jatropha curcas] |
| 5 |
Hb_000244_260 |
0.1077397375 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000254_070 |
0.1079749708 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 7 |
Hb_002367_110 |
0.1106480367 |
- |
- |
PREDICTED: cinnamyl alcohol dehydrogenase 1 [Sesamum indicum] |
| 8 |
Hb_000000_120 |
0.1132605437 |
- |
- |
hypothetical protein EUGRSUZ_A004731, partial [Eucalyptus grandis] |
| 9 |
Hb_000029_440 |
0.1138462453 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit isoform X2 [Vitis vinifera] |
| 10 |
Hb_000679_040 |
0.1225963813 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 11 |
Hb_000674_010 |
0.1245516622 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_069619_010 |
0.1251372729 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 13 |
Hb_003058_200 |
0.1289869505 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 14 |
Hb_028077_020 |
0.1306618746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000019_170 |
0.1306951048 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 16 |
Hb_001105_130 |
0.1313485967 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 17 |
Hb_000220_080 |
0.1322422587 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_001006_270 |
0.1322940677 |
- |
- |
peptide transporter, putative [Ricinus communis] |
| 19 |
Hb_009535_030 |
0.1354878826 |
- |
- |
CP2 [Hevea brasiliensis] |
| 20 |
Hb_000701_030 |
0.1369910166 |
- |
- |
Sodium/hydrogen exchanger 6 -like protein [Gossypium arboreum] |