| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
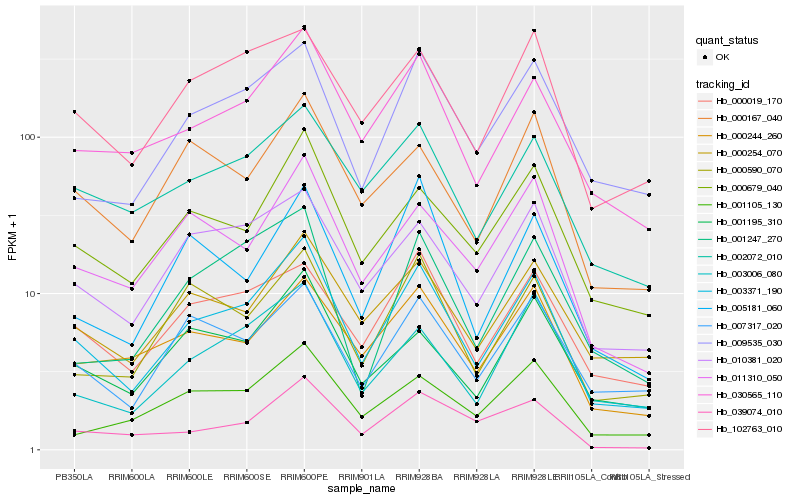
Hb_003371_190 |
0.0 |
- |
- |
PREDICTED: beta-glucosidase 13-like [Jatropha curcas] |
| 2 |
Hb_030565_110 |
0.0952537718 |
- |
- |
PREDICTED: adenosylhomocysteinase 1 [Jatropha curcas] |
| 3 |
Hb_000679_040 |
0.1090074733 |
- |
- |
Cellulose synthase 1 [Theobroma cacao] |
| 4 |
Hb_002072_010 |
0.1151088029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010381_020 |
0.1161588877 |
- |
- |
Endosomal P24A protein precursor, putative [Ricinus communis] |
| 6 |
Hb_009535_030 |
0.1189456107 |
- |
- |
CP2 [Hevea brasiliensis] |
| 7 |
Hb_000254_070 |
0.1204839699 |
- |
- |
PREDICTED: protein disulfide isomerase-like 1-6 [Jatropha curcas] |
| 8 |
Hb_011310_050 |
0.123334118 |
- |
- |
PREDICTED: cellulose synthase A catalytic subunit 1 [UDP-forming] [Jatropha curcas] |
| 9 |
Hb_102763_010 |
0.1254204888 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 10 |
Hb_007317_020 |
0.1257099373 |
- |
- |
PREDICTED: transmembrane 9 superfamily member 5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000019_170 |
0.1258036264 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 12 |
Hb_003006_080 |
0.1261066512 |
- |
- |
PREDICTED: heparan-alpha-glucosaminide N-acetyltransferase-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000590_070 |
0.1262681689 |
- |
- |
PREDICTED: beta-hexosaminidase 1 [Jatropha curcas] |
| 14 |
Hb_000244_260 |
0.1271196528 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001195_310 |
0.1274222804 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_039074_010 |
0.1294254575 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_000167_040 |
0.1312137204 |
- |
- |
PREDICTED: serine carboxypeptidase-like [Jatropha curcas] |
| 18 |
Hb_001105_130 |
0.1325799445 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 19 |
Hb_001247_270 |
0.1381290541 |
- |
- |
PREDICTED: probable L-cysteine desulfhydrase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005181_060 |
0.1391419227 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |