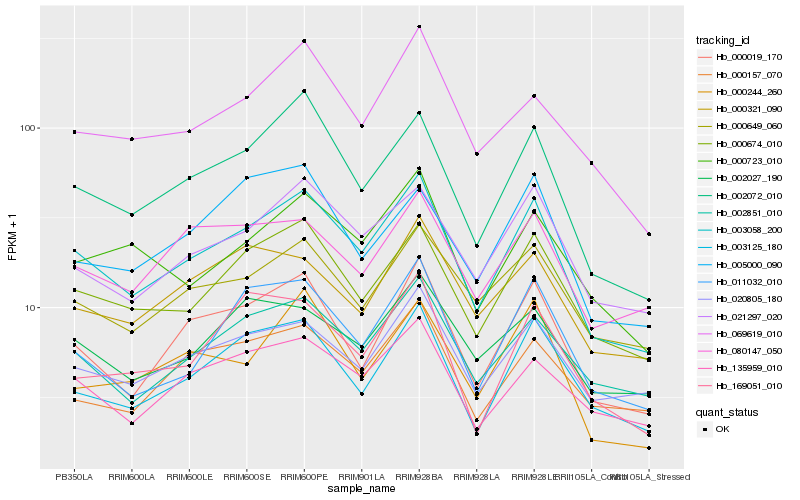
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003058_200 |
0.0 |
- |
- |
PREDICTED: dihydropyrimidinase [Populus euphratica] |
| 2 |
Hb_002072_010 |
0.0758968731 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000674_010 |
0.0769360795 |
- |
- |
PREDICTED: copper-transporting ATPase RAN1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000019_170 |
0.0804900678 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 5 |
Hb_021297_020 |
0.0859939102 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 6 |
Hb_002851_010 |
0.0862550544 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 7 |
Hb_003125_180 |
0.0904973632 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 8 |
Hb_020805_180 |
0.0935488932 |
- |
- |
PREDICTED: epidermal growth factor receptor substrate 15-like 1 [Jatropha curcas] |
| 9 |
Hb_135959_010 |
0.0940292267 |
- |
- |
hypothetical protein JCGZ_07060 [Jatropha curcas] |
| 10 |
Hb_011032_010 |
0.0946324144 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 11 |
Hb_000649_060 |
0.0987766984 |
- |
- |
PREDICTED: E3 ubiquitin protein ligase RIE1 [Jatropha curcas] |
| 12 |
Hb_069619_010 |
0.1021798293 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 13 |
Hb_000723_010 |
0.1042720909 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 14 |
Hb_002027_190 |
0.1061089154 |
- |
- |
PREDICTED: autophagy-related protein 18g [Jatropha curcas] |
| 15 |
Hb_000157_070 |
0.1080393259 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 16 |
Hb_000244_260 |
0.1123743224 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |
| 17 |
Hb_080147_050 |
0.1126386392 |
- |
- |
PREDICTED: probable plastidic glucose transporter 2 [Jatropha curcas] |
| 18 |
Hb_000321_090 |
0.1126645106 |
- |
- |
PREDICTED: calcium-transporting ATPase 1, endoplasmic reticulum-type-like [Jatropha curcas] |
| 19 |
Hb_169051_010 |
0.1177888157 |
- |
- |
JHL06B08.1 [Jatropha curcas] |
| 20 |
Hb_005000_090 |
0.1184112482 |
- |
- |
PREDICTED: venom phosphodiesterase 2 [Jatropha curcas] |