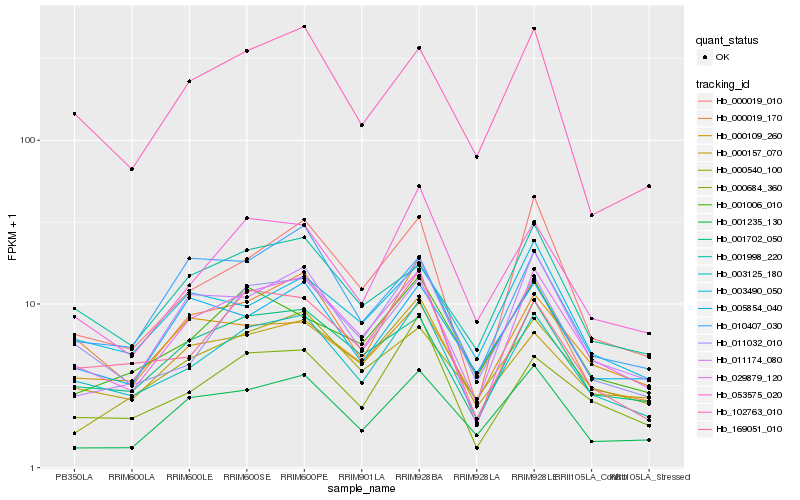
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029879_120 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001702_050 |
0.0723517534 |
- |
- |
PREDICTED: probable protein S-acyltransferase 19 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003125_180 |
0.0785081353 |
- |
- |
hypothetical protein JCGZ_26440 [Jatropha curcas] |
| 4 |
Hb_000019_010 |
0.0903765497 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Jatropha curcas] |
| 5 |
Hb_011174_080 |
0.0979183687 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 6 |
Hb_001235_130 |
0.1110724886 |
- |
- |
- |
| 7 |
Hb_000157_070 |
0.111999265 |
- |
- |
PREDICTED: alpha-N-acetylglucosaminidase [Jatropha curcas] |
| 8 |
Hb_000019_170 |
0.1142234205 |
- |
- |
PREDICTED: alpha-mannosidase [Jatropha curcas] |
| 9 |
Hb_000684_360 |
0.1148376307 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_16454 [Jatropha curcas] |
| 10 |
Hb_001998_220 |
0.115845366 |
- |
- |
PREDICTED: isoamylase 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000109_260 |
0.118382978 |
- |
- |
PREDICTED: WAT1-related protein At3g28050-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_169051_010 |
0.1184888004 |
- |
- |
JHL06B08.1 [Jatropha curcas] |
| 13 |
Hb_011032_010 |
0.1200984314 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 14 |
Hb_010407_030 |
0.1207946323 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 15 |
Hb_005854_040 |
0.1234203934 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_003490_050 |
0.1241452443 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic [Jatropha curcas] |
| 17 |
Hb_102763_010 |
0.1253060187 |
- |
- |
PREDICTED: heat shock cognate protein 80 [Jatropha curcas] |
| 18 |
Hb_053575_020 |
0.125970041 |
- |
- |
PREDICTED: RNA-binding protein 39 [Jatropha curcas] |
| 19 |
Hb_000540_100 |
0.1263984134 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 20 |
Hb_001006_010 |
0.1313917683 |
- |
- |
PREDICTED: putative nuclease HARBI1 [Jatropha curcas] |