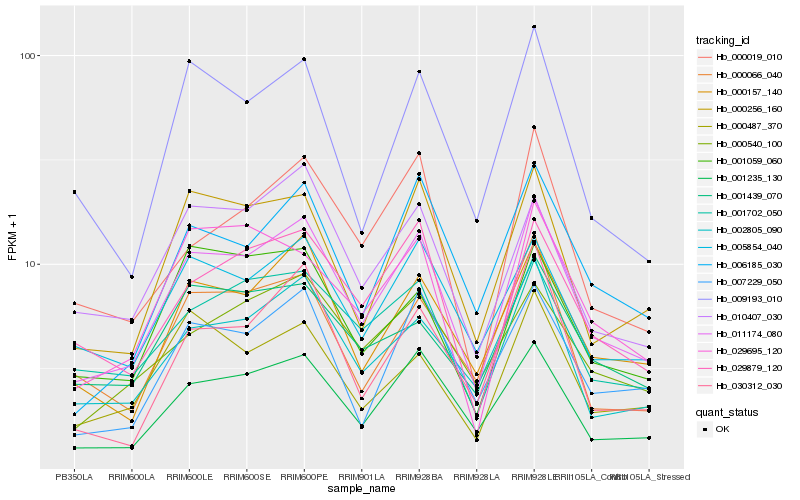
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011174_080 |
0.0 |
- |
- |
PREDICTED: sulfite oxidase [Jatropha curcas] |
| 2 |
Hb_001235_130 |
0.0783099438 |
- |
- |
- |
| 3 |
Hb_000157_140 |
0.0934197885 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_002805_090 |
0.0967799503 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 5 |
Hb_000256_160 |
0.0973053088 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 6 |
Hb_029879_120 |
0.0979183687 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001702_050 |
0.1035570833 |
- |
- |
PREDICTED: probable protein S-acyltransferase 19 isoform X1 [Jatropha curcas] |
| 8 |
Hb_005854_040 |
0.1043002664 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_006185_030 |
0.1062911679 |
- |
- |
threonine synthase, putative [Ricinus communis] |
| 10 |
Hb_007229_050 |
0.1064147817 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_010407_030 |
0.1093692001 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 12 |
Hb_009193_010 |
0.1119360834 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 13 |
Hb_000019_010 |
0.1119416518 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Jatropha curcas] |
| 14 |
Hb_000066_040 |
0.1136124155 |
- |
- |
PREDICTED: CDPK-related kinase 7-like [Populus euphratica] |
| 15 |
Hb_000487_370 |
0.1136714982 |
- |
- |
- |
| 16 |
Hb_000540_100 |
0.1142086152 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g60050 [Jatropha curcas] |
| 17 |
Hb_029695_120 |
0.1165019419 |
- |
- |
hypothetical protein POPTR_0002s05320g [Populus trichocarpa] |
| 18 |
Hb_001439_070 |
0.1167379125 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 19 |
Hb_030312_030 |
0.1169524399 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP40 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001059_060 |
0.1170203615 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |