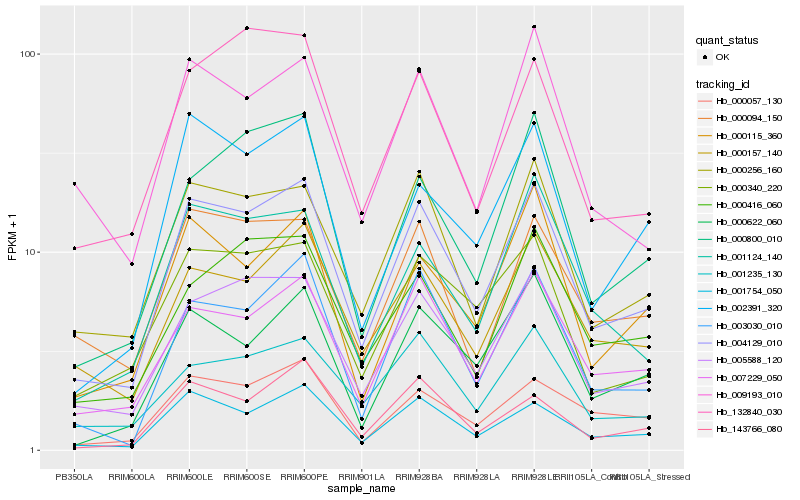
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004129_010 |
0.0 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 2 |
Hb_005588_120 |
0.0749372905 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001754_050 |
0.0775152935 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_007229_050 |
0.083217975 |
- |
- |
PREDICTED: palmitoyl-protein thioesterase 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000256_160 |
0.0881178143 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein knotted-1-like 3-like isoform X1 [Glycine max] |
| 6 |
Hb_000157_140 |
0.0963874933 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 7 |
Hb_003030_010 |
0.0968065736 |
- |
- |
PREDICTED: random slug protein 5-like [Jatropha curcas] |
| 8 |
Hb_000340_220 |
0.1085800493 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001235_130 |
0.1090761356 |
- |
- |
- |
| 10 |
Hb_000057_130 |
0.1094747444 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 11 |
Hb_000094_150 |
0.1113175728 |
- |
- |
- |
| 12 |
Hb_001124_140 |
0.1113268429 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 13 |
Hb_143766_080 |
0.114279569 |
- |
- |
PREDICTED: uncharacterized protein LOC105637797 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000622_060 |
0.1148975884 |
- |
- |
PREDICTED: uncharacterized protein At5g05190-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_002391_320 |
0.1152588355 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 16 |
Hb_000416_060 |
0.1157048029 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 17 |
Hb_000115_360 |
0.1169688306 |
- |
- |
PREDICTED: probable protein phosphatase 2C 15 isoform X3 [Jatropha curcas] |
| 18 |
Hb_000800_010 |
0.1172448236 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 19 |
Hb_009193_010 |
0.1176736249 |
- |
- |
PREDICTED: PLASMODESMATA CALLOSE-BINDING PROTEIN 3 [Jatropha curcas] |
| 20 |
Hb_132840_030 |
0.1201207086 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |