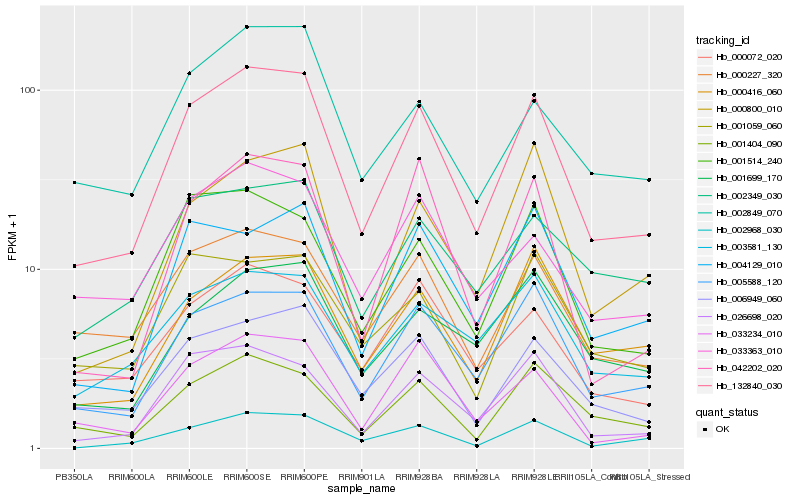
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132840_030 |
0.0 |
transcription factor |
TF Family: MYB-related |
lhy1 [Populus tremula] |
| 2 |
Hb_005588_120 |
0.085959422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006949_060 |
0.0943293851 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 4 |
Hb_001514_240 |
0.10238029 |
- |
- |
PREDICTED: aspartic proteinase CDR1 [Jatropha curcas] |
| 5 |
Hb_026698_020 |
0.1062595775 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 6 |
Hb_000227_320 |
0.1078263536 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 7 |
Hb_001404_090 |
0.1110816676 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 8 |
Hb_000416_060 |
0.1131848351 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 9 |
Hb_000072_020 |
0.1134847424 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 10 |
Hb_002968_030 |
0.1161047752 |
- |
- |
Pectinesterase-1 precursor, putative [Ricinus communis] |
| 11 |
Hb_033234_010 |
0.1187621723 |
desease resistance |
Gene Name: LRR_8 |
Disease resistance protein RPP8 [Theobroma cacao] |
| 12 |
Hb_003581_130 |
0.1195899264 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_004129_010 |
0.1201207086 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 14 |
Hb_000800_010 |
0.1202435269 |
- |
- |
PREDICTED: glycosyltransferase family protein 64 protein C5 [Jatropha curcas] |
| 15 |
Hb_033363_010 |
0.120853343 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 16 |
Hb_002849_070 |
0.1218369636 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_042202_020 |
0.1228807319 |
- |
- |
Eukaryotic release factor 1-3 isoform 1 [Theobroma cacao] |
| 18 |
Hb_001699_170 |
0.1259910628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002349_030 |
0.1280130197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001059_060 |
0.1285728476 |
- |
- |
PREDICTED: bifunctional riboflavin kinase/FMN phosphatase [Jatropha curcas] |