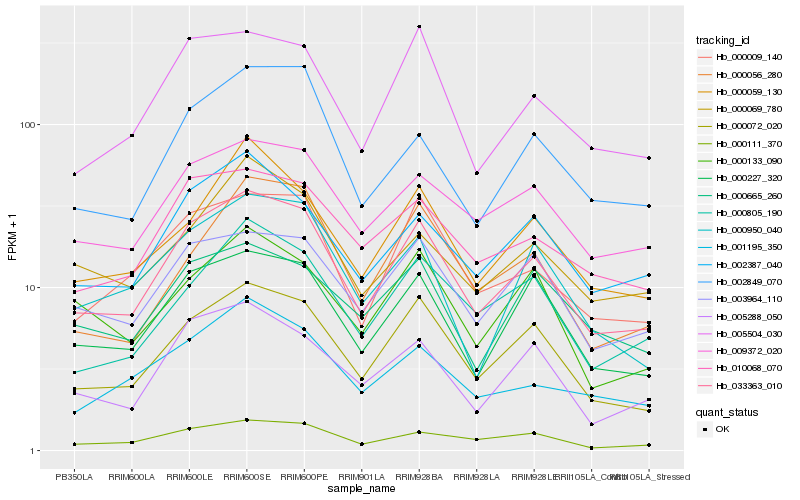
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033363_010 |
0.0 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 2 |
Hb_000072_020 |
0.0650845397 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000227_320 |
0.0842836754 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_000009_140 |
0.0904211963 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 5 |
Hb_000111_370 |
0.0904445774 |
- |
- |
PREDICTED: cation/H(+) antiporter 2-like [Jatropha curcas] |
| 6 |
Hb_009372_020 |
0.0914450174 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit alpha-like [Jatropha curcas] |
| 7 |
Hb_005504_030 |
0.0936018183 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 8 |
Hb_010068_070 |
0.0949552368 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 9 |
Hb_003964_110 |
0.0955757354 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002387_040 |
0.0960047655 |
- |
- |
protein transporter, putative [Ricinus communis] |
| 11 |
Hb_005288_050 |
0.0973141561 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR4 [Jatropha curcas] |
| 12 |
Hb_000805_190 |
0.1049440067 |
transcription factor |
TF Family: GRAS |
PREDICTED: uncharacterized protein LOC103323607 [Prunus mume] |
| 13 |
Hb_000059_130 |
0.1054297679 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 14 |
Hb_002849_070 |
0.106439184 |
- |
- |
PREDICTED: protein BPS1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000950_040 |
0.1075798717 |
- |
- |
cation-transporting atpase plant, putative [Ricinus communis] |
| 16 |
Hb_001195_350 |
0.1084619208 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |
| 17 |
Hb_000056_280 |
0.11593541 |
- |
- |
sur2 hydroxylase/desaturase, putative [Ricinus communis] |
| 18 |
Hb_000069_780 |
0.116143778 |
- |
- |
atpob1, putative [Ricinus communis] |
| 19 |
Hb_000133_090 |
0.116614767 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 20 |
Hb_000665_260 |
0.1167815685 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |