| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
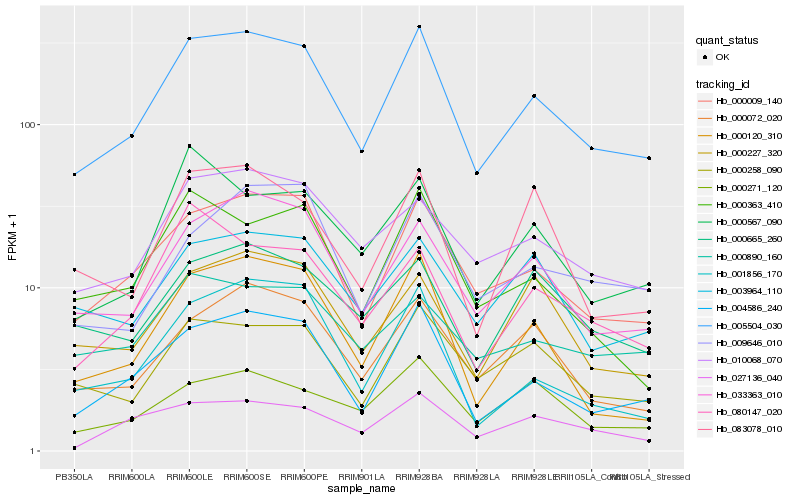
Hb_005504_030 |
0.0 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 2 |
Hb_000009_140 |
0.0790974569 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 3 |
Hb_004586_240 |
0.0810927144 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 4 |
Hb_033363_010 |
0.0936018183 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 5 |
Hb_000258_090 |
0.1085312202 |
- |
- |
PREDICTED: KH domain-containing protein At4g18375 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000072_020 |
0.110280595 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 7 |
Hb_010068_070 |
0.1123269251 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 8 |
Hb_000120_310 |
0.1202269446 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 9 |
Hb_003964_110 |
0.1211515194 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |
| 10 |
Hb_000227_320 |
0.1220460694 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 11 |
Hb_000665_260 |
0.1262239241 |
- |
- |
PREDICTED: uncharacterized protein LOC105637620 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000271_120 |
0.1262951662 |
- |
- |
PREDICTED: U-box domain-containing protein 13-like [Nelumbo nucifera] |
| 13 |
Hb_000363_410 |
0.1275329194 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 14 |
Hb_027136_040 |
0.127884387 |
- |
- |
hypothetical protein JCGZ_00277 [Jatropha curcas] |
| 15 |
Hb_009646_010 |
0.1285364836 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 16 |
Hb_000890_160 |
0.1289379724 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 17 |
Hb_000567_090 |
0.1290398539 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_080147_020 |
0.1292632054 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 19 |
Hb_001856_170 |
0.129698558 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 20 |
Hb_083078_010 |
0.1298699817 |
- |
- |
Splicing factor, arginine/serine-rich, putative [Ricinus communis] |