| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
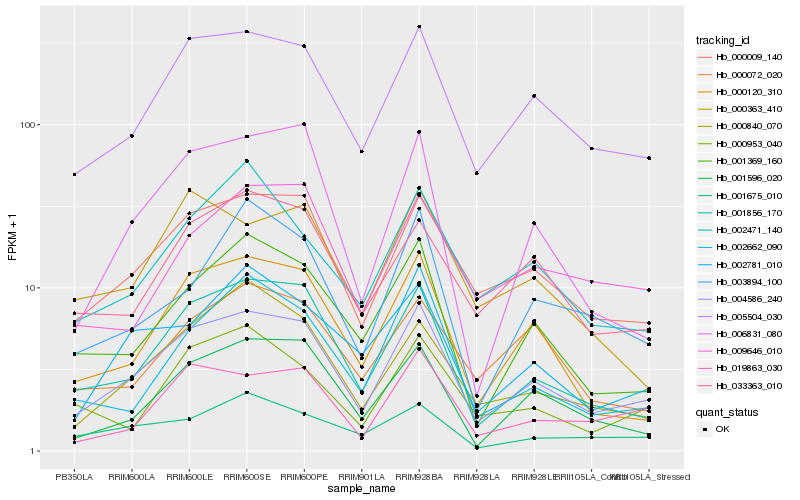
Hb_004586_240 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 2 |
Hb_005504_030 |
0.0810927144 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 3 |
Hb_000009_140 |
0.0884616743 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 4 |
Hb_001856_170 |
0.1015886264 |
- |
- |
PREDICTED: ABC transporter B family member 27 [Jatropha curcas] |
| 5 |
Hb_000120_310 |
0.1159595852 |
- |
- |
Gibberellin 3-beta-dioxygenase, putative [Ricinus communis] |
| 6 |
Hb_001369_160 |
0.1262945182 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 7 |
Hb_001596_020 |
0.1268237654 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 8 |
Hb_006831_080 |
0.1285302594 |
- |
- |
PREDICTED: uncharacterized protein LOC105642126 [Jatropha curcas] |
| 9 |
Hb_019863_030 |
0.129571183 |
- |
- |
hypothetical protein JCGZ_00393 [Jatropha curcas] |
| 10 |
Hb_033363_010 |
0.1328525121 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 11 |
Hb_000840_070 |
0.1336364296 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_002781_010 |
0.1375491627 |
transcription factor |
TF Family: LUG |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_001675_010 |
0.1403295657 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 14 |
Hb_009646_010 |
0.1474504464 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 15 |
Hb_002471_140 |
0.1480675695 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 16 |
Hb_003894_100 |
0.150433066 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 17 |
Hb_000953_040 |
0.1507159254 |
- |
- |
hypothetical protein OsJ_19135 [Oryza sativa Japonica Group] |
| 18 |
Hb_000363_410 |
0.1510413431 |
- |
- |
hypothetical protein JCGZ_00955 [Jatropha curcas] |
| 19 |
Hb_002662_090 |
0.1512774607 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 20 |
Hb_000072_020 |
0.1517616596 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |