| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
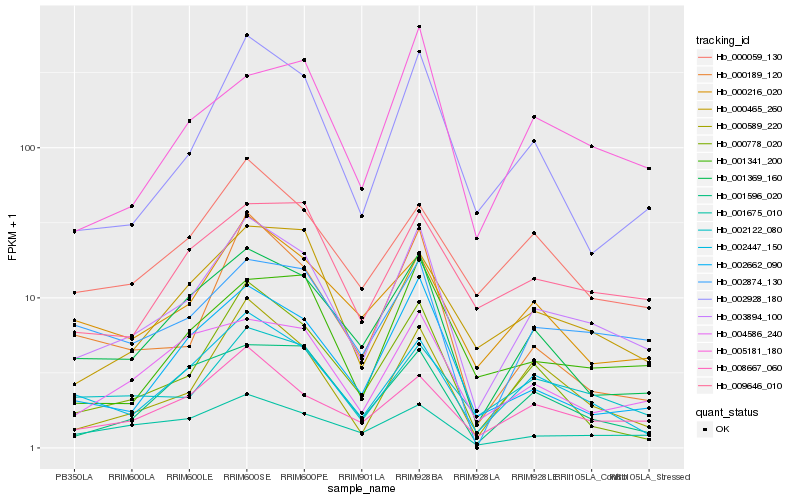
Hb_003894_100 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 2 |
Hb_001369_160 |
0.1261126123 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 3 |
Hb_002447_150 |
0.1268834521 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 4 |
Hb_008667_060 |
0.1284915604 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 5 |
Hb_000589_220 |
0.1293683751 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: uncharacterized protein LOC105124982 [Populus euphratica] |
| 6 |
Hb_002928_180 |
0.1378215745 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 7 |
Hb_002122_080 |
0.1380545881 |
- |
- |
- |
| 8 |
Hb_002662_090 |
0.138673186 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 9 |
Hb_000778_020 |
0.1461986605 |
- |
- |
PREDICTED: salicylic acid-binding protein 2-like [Jatropha curcas] |
| 10 |
Hb_002874_130 |
0.1466507291 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 11 |
Hb_000465_260 |
0.1500020896 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT4-like [Jatropha curcas] |
| 12 |
Hb_001675_010 |
0.1502805009 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 13 |
Hb_001596_020 |
0.1503557976 |
- |
- |
hypothetical protein CICLE_v10014140mg [Citrus clementina] |
| 14 |
Hb_004586_240 |
0.150433066 |
- |
- |
PREDICTED: serine/threonine-protein kinase-like protein At1g28390 [Jatropha curcas] |
| 15 |
Hb_000216_020 |
0.1516575349 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 16 |
Hb_001341_200 |
0.1519287147 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 4 [Jatropha curcas] |
| 17 |
Hb_000059_130 |
0.1544090874 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 18 |
Hb_009646_010 |
0.1551011874 |
- |
- |
PREDICTED: hyoscyamine 6-dioxygenase-like [Jatropha curcas] |
| 19 |
Hb_000189_120 |
0.1559681113 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 20 |
Hb_005181_180 |
0.1586547505 |
- |
- |
conserved hypothetical protein [Ricinus communis] |