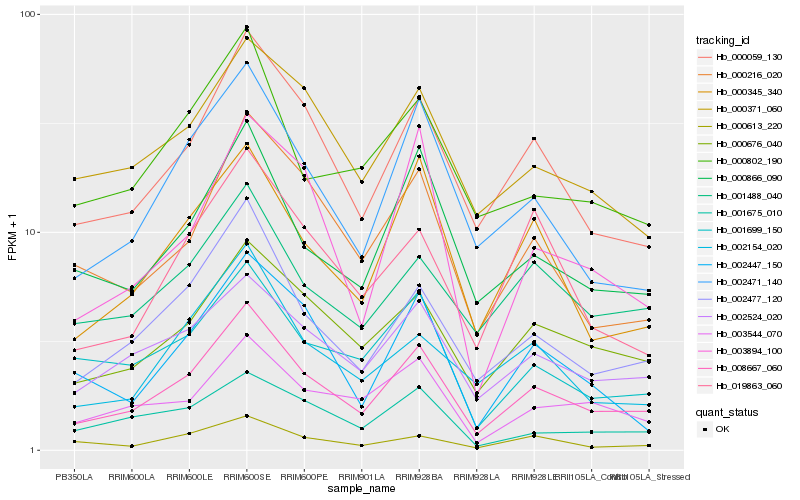
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008667_060 |
0.0 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 2 |
Hb_000059_130 |
0.0882107462 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 3 |
Hb_002477_120 |
0.0912411203 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 4 |
Hb_000676_040 |
0.0994074505 |
- |
- |
hypothetical protein POPTR_0007s13320g [Populus trichocarpa] |
| 5 |
Hb_002471_140 |
0.1026300317 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 6 |
Hb_002524_020 |
0.1079297915 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 7 |
Hb_000345_340 |
0.1085423236 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 8 |
Hb_000866_090 |
0.1099136578 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_000216_020 |
0.1177389057 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 10 |
Hb_003544_070 |
0.1233688041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001699_150 |
0.1234840241 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 12 |
Hb_000371_060 |
0.1274619955 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 13 |
Hb_003894_100 |
0.1284915604 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 1-like [Jatropha curcas] |
| 14 |
Hb_002154_020 |
0.1289190146 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_001488_040 |
0.1295133893 |
- |
- |
PREDICTED: nipped-B-like protein A [Jatropha curcas] |
| 16 |
Hb_001675_010 |
0.1305347453 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 17 |
Hb_019863_060 |
0.1313301259 |
- |
- |
PREDICTED: target of Myb protein 1-like [Populus euphratica] |
| 18 |
Hb_000802_190 |
0.1329688349 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 19 |
Hb_000613_220 |
0.1332515062 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 20 |
Hb_002447_150 |
0.1343627703 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |