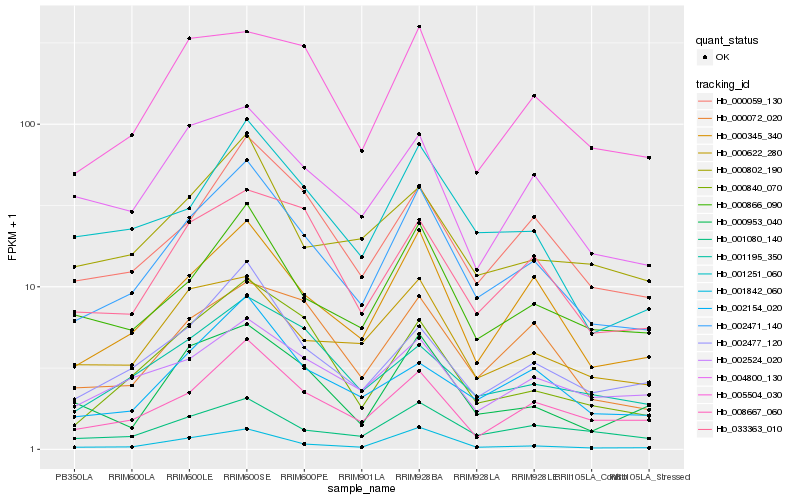
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002471_140 |
0.0 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 2 |
Hb_000345_340 |
0.0878711011 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 3 |
Hb_008667_060 |
0.1026300317 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 4 |
Hb_000059_130 |
0.1049792143 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 5 |
Hb_000866_090 |
0.1079325467 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_001251_060 |
0.1139848575 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 7 |
Hb_002477_120 |
0.114247797 |
- |
- |
PREDICTED: phospholipid:diacylglycerol acyltransferase 1-like [Jatropha curcas] |
| 8 |
Hb_001080_140 |
0.122614983 |
transcription factor |
TF Family: RWP-RK |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000840_070 |
0.1233819815 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 10 |
Hb_000072_020 |
0.1266748579 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 11 |
Hb_033363_010 |
0.1267039237 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 8 [Jatropha curcas] |
| 12 |
Hb_002154_020 |
0.1271421925 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_001842_060 |
0.1287784139 |
desease resistance |
Gene Name: ABC_trans_N |
hypothetical protein JCGZ_10908 [Jatropha curcas] |
| 14 |
Hb_000953_040 |
0.1299346193 |
- |
- |
hypothetical protein OsJ_19135 [Oryza sativa Japonica Group] |
| 15 |
Hb_000622_280 |
0.1323765723 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004800_130 |
0.1330533944 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 17 |
Hb_005504_030 |
0.1340404968 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 2-like [Pyrus x bretschneideri] |
| 18 |
Hb_002524_020 |
0.1354610393 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 19 |
Hb_000802_190 |
0.1363719125 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 20 |
Hb_001195_350 |
0.1370378994 |
- |
- |
hypothetical protein JCGZ_04423 [Jatropha curcas] |