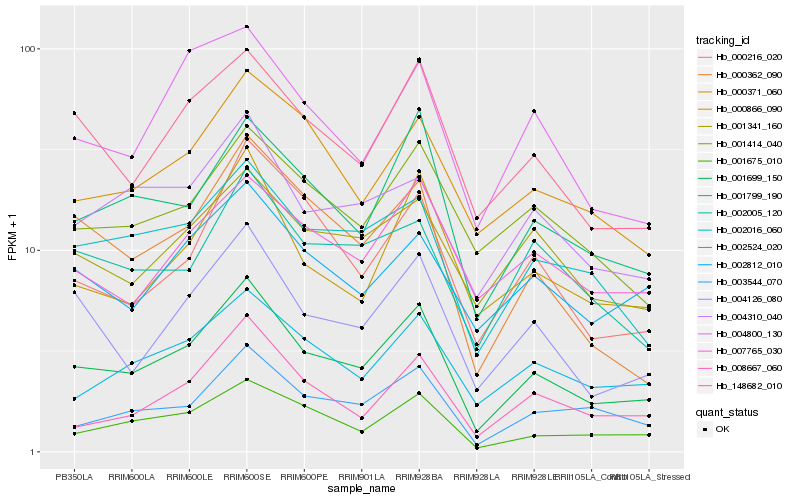
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001699_150 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 2 |
Hb_001799_190 |
0.0986663563 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 3 |
Hb_004126_080 |
0.1011186164 |
- |
- |
mitochondrial 50S ribosomal protein L21 [Hevea brasiliensis] |
| 4 |
Hb_148682_010 |
0.1026557373 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 5 |
Hb_001675_010 |
0.102996166 |
- |
- |
hypothetical protein TRIUR3_09961 [Triticum urartu] |
| 6 |
Hb_002016_060 |
0.1077648523 |
- |
- |
PREDICTED: uncharacterized protein LOC105642502 [Jatropha curcas] |
| 7 |
Hb_003544_070 |
0.1108499521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000866_090 |
0.1112943118 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 9 |
Hb_002524_020 |
0.1171258908 |
- |
- |
glycerophosphoryl diester phosphodiesterase, putative [Ricinus communis] |
| 10 |
Hb_000362_090 |
0.118837657 |
- |
- |
PREDICTED: uncharacterized protein LOC105635736 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004310_040 |
0.120050998 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000216_020 |
0.1218768091 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 13 |
Hb_000371_060 |
0.1221409483 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 14 |
Hb_004800_130 |
0.1226831118 |
- |
- |
hypothetical protein POPTR_0005s12580g [Populus trichocarpa] |
| 15 |
Hb_008667_060 |
0.1234840241 |
- |
- |
Magnesium and cobalt efflux protein corC, putative [Ricinus communis] |
| 16 |
Hb_002812_010 |
0.1240637482 |
- |
- |
PREDICTED: chitin elicitor receptor kinase 1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001414_040 |
0.1242639394 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001341_160 |
0.1244097706 |
- |
- |
PREDICTED: E3 SUMO-protein ligase SIZ1 isoform X2 [Jatropha curcas] |
| 19 |
Hb_007765_030 |
0.1256349146 |
- |
- |
PREDICTED: neutral ceramidase [Jatropha curcas] |
| 20 |
Hb_002005_120 |
0.1268809604 |
- |
- |
PREDICTED: protein MEI2-like 5 [Jatropha curcas] |